“Each of us is on our own trajectory – steered by our genes and our experiences – and as a result every brain has a different internal life. Brains are as unique as snowflakes.”
David Eagleman
Introduction
The rapidly expanding wavefront of Generative AI (Gen AI), in the last couple of years, can be largely attributed to a seminal paper “Attention is all you need” by Google. This paper by Google researchers, was a landmark in the field of AI and led to an explosion of ideas and spawned an entire industry based on its theme. The paper introduces 2 key concepts a) Attention mechanism the b) Transformer Architecture. In this post, I distil the essence of Attention and Transformers.
Transformers were originally invented for Natural Language Processing (NLP) tasks like language translation, classification, sentiment analysis, summarisation, chat sessions etc. However, it led to its adaptation to languages, voice, music, images, videos etc. Prior to the advent of transformers, Natural Language Processing (NLP) was largely done through Recurrent Neural Networks (RNNs) . The problem with encoder-decoder based RNNs is that it had a fixed length for an internal-hidden state, which stored the information, for translation or other NLP tasks. Clearly, it was difficult to capture all the relevant information in this fixed length internal-hidden state. A single, final hidden state had to capture all information from the input sequence enabling it to generate the output sequence for translation and other tasks. There were some enhancements to address the shortcomings of RNNs with approaches such as Long Short-term Memory(LSTM), Gated Recurrent Unit (GRU) etc., but by and large the performance of these NLP models fell short of being reliable and consistent. This shortcoming was addressed in the paper by Bahdanau et al in the paper ‘Neural machine translation by jointly learning to align and translate‘, which discussed how ‘attention’ can be used to identify which words, align to which other words in its neighbourhood, which is computed as context vector. It implemented a ‘mechanism of attention’ by enabling the decoder to decide which parts of the sentence it needs to pay attention to, thus relieving the encoder to encode all information of the sentence into a single internal-hidden state
The attention-based transformer architecture in the paper ‘Attention is all you need‘ took its inspiration from the above Bahdanau paper and eventually evolved into the current Large Language Models (LLMs). The transformer architecture based on the attention mechanism was able to effectively address the shortcomings of the earlier RNNs. The architecture of the LLM is based on 2 key principles
a. An attention mechanism which determines the relationships between words in a sequence. It identifies how each word relates to others words in the sequence
b. A feed-forward neural network that takes the output of the attention module and enriches the relationships between the words
A final layer using softmax can predict the next word in a given sequence
LLM’s are based on the Transformer architecture. LLMs like ChatGPT, Claude, Cohere, Llama etc., typically go through 2 stages a) Pre-training b) Fine tuning
During pre-training the LLM is trained on a large corpus of unstructured text from the internet, wikipedia, arXiv, stack overflow etc. The pre-training helps the LLMs in general language understanding, enabling LLMs to learn grammar, syntax, context etc. This is followed by a fine-tuning phase where the language models is trained for specific domain or task using a smaller curated and annotated dataset of input, output pairs. This adjusts the weights of the LLM to handle the specific task in a particular domain. This may be further enhanced based on Reinforcement Learning with Human Feedback (RLHF) with reward-penalty for a given task during training. (In many ways, we humans also go through the stages of pre-training and fine-tuning in my opinion. As David Eagleman states above, we all come with a genetic blueprint based on millions of years of evolution of responses to triggers. During our early formative years this genetic DNA will create certain neural pathways in the brain akin to pre-training. Further from 2-5 years, through a couple of years of fine-tuning we learn a lot more – languages, recognition, emotion etc. This does simplify things to an extent but still I think to a large extent it holds)
Clearly, our brain is not only much more complex but also uses a minuscule energy about 60W to compute complex tasks, which is roughly equivalent to a light bulb. While for e.g. training GPT-3 which has 175 billion parameters, consumes an estimated 1287 MWH, which is roughly equivalent the consumption of an average US household for 120 years (Ref: https://adasci.org/how-much-energy-do-llms-consume-unveiling-the-power-behind-ai/?ts=1734961973)
NLP is based on the fact that human language is some ordered sequence of words. Moreover, words in a language are repetitive and thus inherently categorical. Hence, we need to use a technique for handling these categorical words for e.g. One-Hot-Encoding (OHE). But since the vocabulary of languages is extremely large, using OHE would become unmanageable. There are several other more efficient encoding methods available. Large Language Models (LLMs), which are the backbone of GenAI are trained on a large corpus of text spanning the internet, wikipedia, and other domains. The text is first converted into a numerical form through a process called tokenisation, where the words, subwords are assigned a numerical value based on some scheme. Tokenisation, can be at the character level, sub-word level, word level, sentence or even paragraph level. The choice of encoding is trade-off between vocabulary size vs sequence or context length. For character level encoding, the vocabulary will be around ~36 including letters, punctuation etc., but the sequences generated for sentences with this method will be very long. While word encodings will have a large vocabulary, an entire sentences can be captured in a shorter sequences. The encodings typically used are Byte Pair Encoding (BPE) from OpenAI, WordPiece or Sentence encoding. The sentences are first converted to tokens.
The tokens are then converted into embedding vectors which can 16, 32 or 128 real-valued dimensions. The embedding vectors convert the tokens into a multi-dimensional continuous space and capture the semantic meaning of the tokens as they get trained. The embeddings assigned, do not inherently capture the semantic meaning of words fully. But in a rough sort of way. For e.g. “I sat on the bank of a river” and “I deposited money in a bank”, the word bank may have the same embedding. But as the model is trained through the transformer with sequences of text passing through the attention module, the embeddings are updated with contextual information. So for e.g. in the 1st sentence “bank” will be associated with the word “river” and in the 2nd sentence the attention module will also capture the context of the “bank” and associate it with the word “money”
A transformer is well suited for predicting the next word in a given sequence. This is called a auto-regressive decoder-only model. The sequence of steps a enable a Transformer to be capable of predicting the next word in a given sequence is based on the following steps
a) Training on a large corpus of text from internet, wikipedia, books, and research repositories like arXiv etc
The text are tokenised based on one of the tokenisation schemes mentioned above like BPE, Wordpiece etc. to convert the words into numerical values
The tokens are then converted into multi-dimensional real-valued embedding vectors. The embeddings are vectors which through multiple iterations capture richer meaning context-aware meaning of sentences
The Attention module determines the affinity each word has to the other words in the sentence. This affinity can be captured over longer sentence structures too and this is based on the context (sequence) length depending on the size of the model.
The weights from the output of the Attention module then go to a simple 2 layer Feed Forward Neural Network (FFN) which tries to predict the next word in a sentence. For this each sentence is taken as input with the target being the same sentence shifted by one place.
For e,g,
Input: Mary had a little lamb
Target: had a little lamb <end>
So in a sentence w1 , w2, w3, … , wn the FFN will use
w1 to predict w2
w1 , w2 to predict w3 and so on
During back propagation, the error between the predicted word and the actual target word is calculated and propagated backwards through the network updating the weights and biases of the NN. The FFN uses tries to minimise the cross-entropy or log loss which computes the difference between the predicted probabilities and target values.
Attention module
For e.g. if we had the sentence “The quick brown fox jumped over the lazy dog”, Attention is computed as follows
Each word in the above sentence is tokenised and represented as dense vector. The transformer architecture uses 3 weight matrices call Wq , Wk, Wv called the Query Weight, Key Weight and Value weight matrices which are learnable matrices. The embedding vectors of the sentence are multiplied with these Wq, Wk, Wv matrices to give Q (Query), K(Key) and V (Value) vectors.
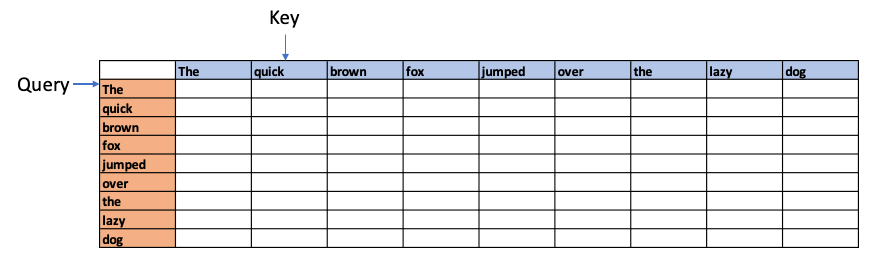
The dot product of the Query vector with all the Key vectors is performed. Since these are vectors, the dot product will determine the similarity or alignment, the query ‘The’ has for the each of the Keys. This is the fundamental concept of of the Attention module. This is because in a multi-dimensional vector space, vectors which are closer together will give a high dot product. Hence, the dot product between the Query and all the Keys gives the affinity the Query has to all other Keys. This is computed as the Attention Score.
For e,g the above process could show that quick and brown attend to the fox, and lazy attends to the dog – and they have relatively high Attention Scores compared to the rest. In addition the Attention operation may also determine that there is a relation between fox and dog in the sentence.

These affinities show up over several iterations through batches of sentences as Wq, WK, Wv are learnable parameters. The relationship learned is depicted below
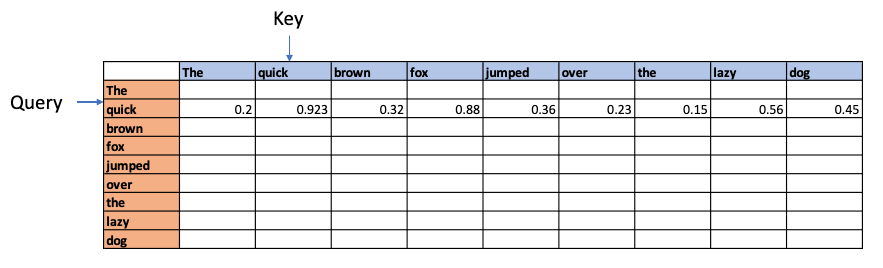
Next the values are normalised using the Softmax function since this will result in a mean of 0 and a variance of 1. This will give normalised attention scores
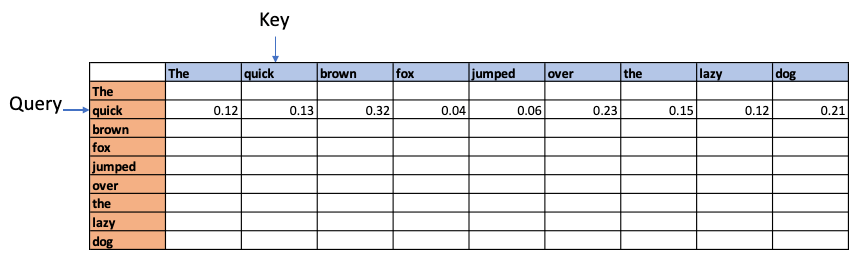
Causal attention. Since future words cannot affect the earlier words these values are made -Infinity so when we perform Softmax we get the value 0 for these values
Self-Attention Mechanism enables the model to evaluate the importance of tokens relative to each other. The self-attention mechanism is written as
where Q, K, V are Query, Key and Value vectors and dK is the dimensionality of the Key vectors. scales the dot product so that the dot product values are not overly large
where the Scaled Attention score =
The Attention weights = softmax(Scaled Attention score)
This computes a context-aware representation of each token with respect to all other tokens
Feed Forward Network (FFN)
In the case of training a language model the fact that language is sequential enables the model to be trained on the language itself. This is done by training the model on large corpus of text, where the language learns to predict the next words in the sequence. So for example in the sentence
Feedforward Network (FFN) comprises two linear transformations separated by a non-linearity, typically modeled
with the first layer transformation as
and the second layer transformation is
where and
are the weight matrices, and
and
are the biases
where x
and
x
and
is usually 4 times the dimesnion of
is the activation function which can be ReLU, GELU or SwiGLU
Input to the FFN
The Decoder receives the output of the Self Attention module to the FFN network. This output from the Attention module is context-aware with the words in the input sequence having different affinities to other words in the sequence

The Target of the FFN is the input sequence shifted by one word

The output from the Attention head and the layer normalization
Normed Output = LayerNorm(Input+MultiHeadOutput)
In essence the Decoder only architecture can be boiled down to the following main modules
- Tokenization – The input text is split either on characters, subwords, words, to convert the text into numbers
- Vector Embedding – The numerical tokens are then converted into Dense vectors
- Positional Embedding – The position order of the text sequence is encoded using the positional embedding. This positional embedding is added to the vector embedding
- Input embedding = Vector embedding + Positional embedding
- Attention module – The attention module computes the affinity the different words have for other words in its vicinity. This is done through the the use of 3 weight matrices
,
,
. By multiplyting these matrices with the input vectors we get Q,K and V vectors. The attention is computed as
- For the decoder, attention is masked to prevent the model from looking at future tokens during training also known as causal attention mentioned above
- The output pf the Attention module is passed to a 2 layer FFN which uses GeLU activation with Adam optimszation. This involves the following 2 steps
- Computing the cross-entropy (loss) using the predicted words and the actual labels
- Backpropagting the error through all the layers and updating the layer weights,
- If the size of the FFN’s output is the vocabulary size then we can use
P(next word|context)=softmax(FFN output)
If the size of the model output is not the vocabulary size then the a final linear layer embeds the output to the size of the dictionary. This maps the model’s hidden states to the vocabulary size enabling the predicting of the next word from the vocabulary - Next word prediction : The next word prediction is done by applying softmax on the output of the FFN layer (logits) to compute the probability for the vocabulary
- P(next word∣context)=softmax(Logits)
- After computing the probability the model selects the next word based on one of many options – to either choose the most probable word or on some other algorithm
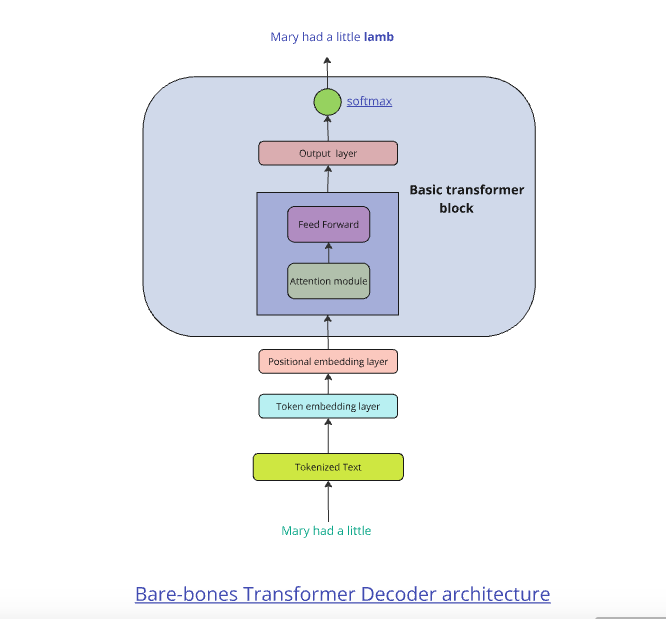
The above sequence of steps is a bare-bones attention and transformer module. In of itself it can achieve little as the transformer module will have to contend with vanishing or exploding gradient issues. It needs additional bells and whistles to make it work effectively
Additional layers to the above architecture
a) Residual Connection and Layer Normalisation (Add + Norm) –
i) Residual, skip connections
Residual connection or skip connections are added the input of each layer to the output to enable the gradients to propagate effectively. This is based on the paper ‘Deep Residual Learning for Image Recognition” from Microsoft
Residual connections also known as skip or shortcut connections are performed by adding the input of layer to the output of the layer as a shortcut. This helps in preventing the vanishing gradient, because of the gradients become progressively smaller as they pass through successive layers.
ii) Layer normalisation
In addition layer normalisation is done to stabilise the activation across a single feature to have 0 mean and a variance of 1 by computing
Mean and variance calculation
,
Normalization
Layer normalization introduces learnable parameters using the equation
This can be written as
ResidualOutput=Input+Output of Attention/FFN
The above statement mentions that the Input layer to the Attention /FFN module is added to the output to mitigate the vanishing gradient problem
NormedOutput=LayerNorm(Residual Output)
Layer Normalisation is then applied to the Residual Output to stabilise the activations.
b) Multi-headed Attention : Typically Transformer use multiple parallel heads of attention. Each head will compute a slightly different variations to the attention values, thus making the whole learning process richer. Multi-headed learning is capable of capturing more nuanced affinities of different words in the sentence to other words in the sentence/context.
c) Dropout : Dropout is a technique where random hidden units or neurons are dropped from the network during training. This prevents overfitting and helps to regularise/generalise the learning of the network. In Transformer Architectures, dropout is used after calculating the Attention Weights. Dropout can also be applied in the Feed Forward Network or in the Residual Connections
This is shown diagrammatically here
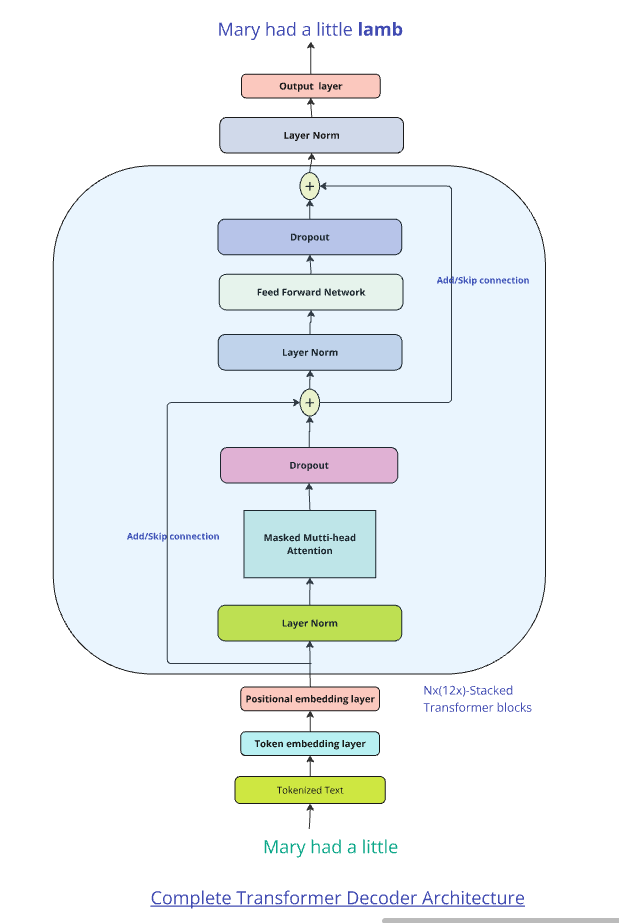
Points to note:
a) The Attention mechanism is able to pick out affinities between words in a sentence. This happens despite the fact the the WQ, WK, WV matrices are randomly initialised. As the model trained iteratively through a large corpus of text using next token prediction for Auto Regressive Transformers and Masked prediction as in the case of BERT, then the affinities show up. This training allows the model to learn the contextual relationships and affinities words have with each other. The dot product Q, K measures the affinity words have for each other and will be high if they are highly related to each other. This is because they will aligned in a the multi-dimensional embedding space of these vectors, besides semantically and contextually related tokens are closer to each other.
b) The Feed Forward Network (FFN) in the Transformer’s Attention block is relatively small and has just 2 layers. This is for computational efficiency and deeper Neural Networks can increase costs. Moreover, it has been found that deeper and wider networks did not significantly improve performance while also preventing overfitting.
c) The above architecture is based on the Causal Attention, Decoder only transformer. The original paper includes both the encoder and the decoder to enable translation across different languages. In addition architectures like BERT use ‘masked attention’ and randomly mask words
The flow of vectors and dimensionality from the input sentence tokens to the output token prediction is as follows
a) For a batch (B) of 2 sentences with 6 words (T) each, where each word is converted into a token. If Byte Pair Encoding (BPE) is used then an integer value between 1-50257 will be obtained.
Input shape = (B x T) = (2 x 6)
b) Token embedding – Each token in the vocabulary is converted into an embedding vector of size = 512 dimension vector
Output shape = (B x T x ) = (2 x 6 x 512)
c) Positional embedding is added
Shape of positional embedding = T x = (6 x512)
d) Output shape with token and positional embedding is the same
Output shape = (B x T x ) = (2 x 6 x 512)
d) Multi-head attention
e) The WQ, WK, WV learnable matrices are each of size
x
f) Q = X x WQ = (B x T x ) x (
x
)
Output shape of Q, K, V = (B x T x ) = (2 x 6 x 512)
g) Number of heads h = 8
Dimensionality of each head = /8 =
= 64
h) Splitting across the heads we have
Shape per head = (B, h, T, ) = ( 2, 8, 6, 64)
h) Weighted sum of values =
Output shape per head = (B, h, T, ) = ( 2, 8, 6, 64)
i) All the heads are concatenated
(B x T x ) = (2 x 6 x 512)
j) The FFN has one hidden layer which is 4 times
=
x 4
Final output of FFN after passing through hidden layer and back
Output shape =(B x T x ) = (2 x 6 x 512)
k) Residual, shortcut connections and layer norm do not change shape
Output shape =(B x T x ) = (2 x 6 x 512)
l) The final output is projected back into the original vocabulary space. For BPE it
50257.
Using a weight matrix (512 x vocab_size) = (512 x 50257)
Final output shape = (B x T x vocab_size) = (2 x 6 x 50257)
The output is in probabilities and hence gives the most likely next word in the sentence
Conclusion
This post tries to condense the key concepts around the Attention mechanism and the Transformer Architecture which have been the catalyst in the last few years, resulting in an explosion in the area of Gen AI, and there seems to be no stopping. It is indeed fascinating how the human language has been mathematically analysed for semantic meaning and relevance.
References
- Building LLMs from Scratch by Sebastian Raschka
- Hands-on Large Language Model by Jay Alammar, Maarten Grootendorst
- Let’s build GPT from scratch: from scratch, in code, spelled out – Andrej Karpathy
- Attention in Transformers, visually explained – 3Blue1Brown
- Awesome LLM – Hannibal046 (collection of LLM papers)
Also see
- Singularity – Short science fiction
- Introducing QCSimulator – A 5 qubit quantum computing simulator in R
- GooglyPlusPlus: Win Probability using Deep Learning and player embeddings
- Natural Language Processing; What would Shakespeare say?
- Deep Learning from first principles in vectorized Python, R and Octave
- Big Data 4: Webserver log analysis with RDDs, Pyspark, SparkR and SparklyR
- Reintroducing cricketr: An R package to analyse performances of cricketers in R
To see all posts, click Index of posts

 Checkout my book ‘Deep Learning from first principles: Second Edition – In vectorized Python, R and Octave’. My book starts with the implementation of a simple 2-layer Neural Network and works its way to a generic L-Layer Deep Learning Network, with all the bells and whistles. The derivations have been discussed in detail. The code has been extensively commented and included in its entirety in the Appendix sections. My book is available on Amazon as
Checkout my book ‘Deep Learning from first principles: Second Edition – In vectorized Python, R and Octave’. My book starts with the implementation of a simple 2-layer Neural Network and works its way to a generic L-Layer Deep Learning Network, with all the bells and whistles. The derivations have been discussed in detail. The code has been extensively commented and included in its entirety in the Appendix sections. My book is available on Amazon as 
 In this book I implement some of the most common, but important Machine Learning algorithms in R and equivalent Python code.
In this book I implement some of the most common, but important Machine Learning algorithms in R and equivalent Python code.
















 Then felt I like some watcher of the skies
Then felt I like some watcher of the skies