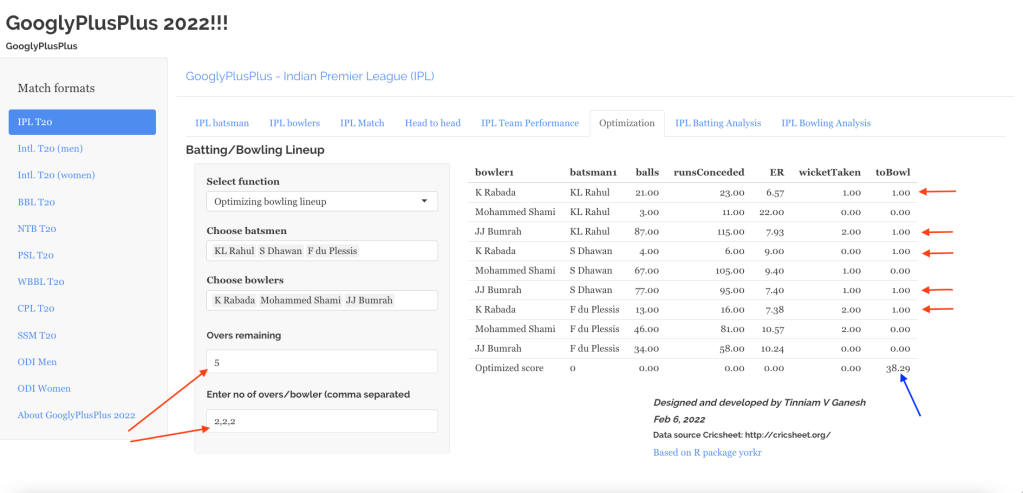
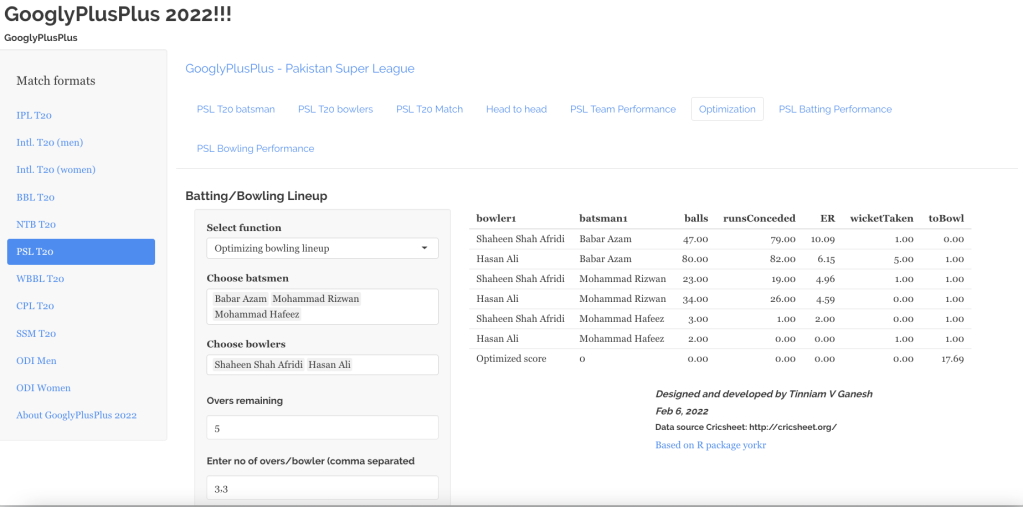
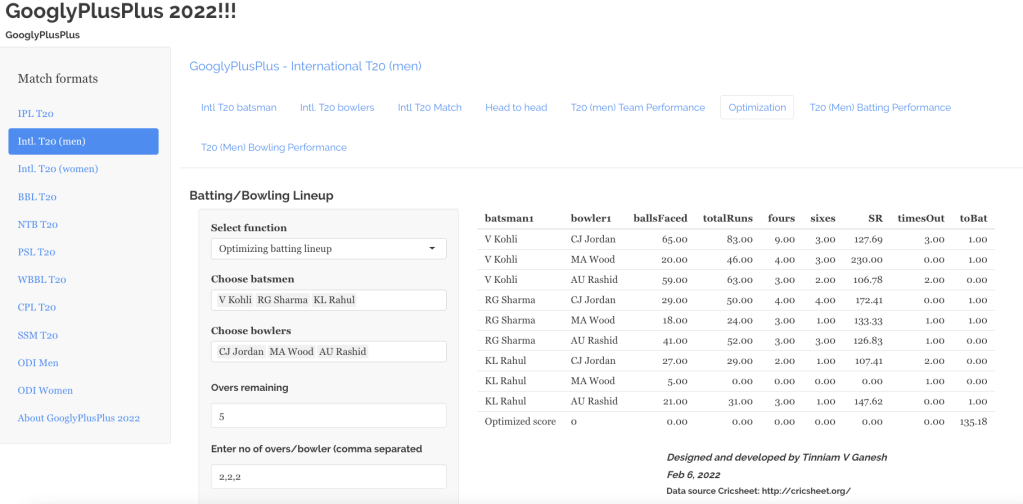
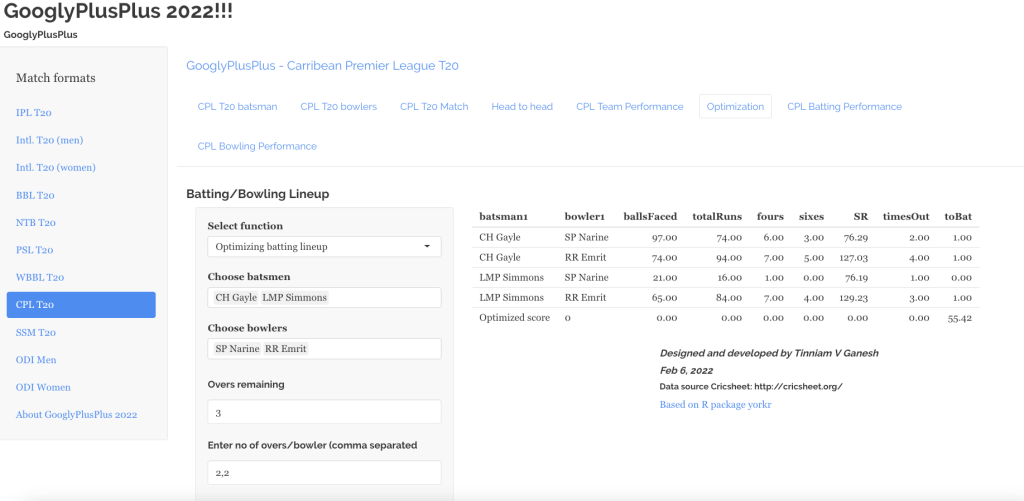
“The unexamined life is not worth living.” – Socrates
“There is no easy way from the earth to the stars.” – Seneca
“If you want to go fast, go alone. If you want to go far, go together.” – African Proverb
1. Introduction
In this post, I put my R package cricketr to analyze the Indian and Australia World Test Championship (WTC) final squad ahead of the World Test Championship 2023.My R package cricketr had its birth on Jul 4, 2015. Cricketr uses data from Cricinfo.
You can download the latest PDF version of the book at ‘Cricket analytics with cricketr and cricpy: Analytics harmony with R and Python-6th edition‘
Indian squad
Rohit Sharma (Captain), Shubman Gill, Cheteshwar Pujara, Virat Kohli, Ajinkya Rahane, Ravindra Jadeja, Shardul Thakur, Mohd. Shami, Mohd. Siraj, Ishan Kishan (wk).
According to me, Ishan Kishan has more experience than KS Bharat, though Rishabh Pant would have been the ideal wicket keeper/left-handed batsman. I think Shardul Thakur would be handful in the English conditions. For a spinner it either Ashwin or Jadeja. Maybe the balance shifts in favor of Jadeja
Australian squad
Pat Cummins (capt), Alex Carey (wk), Cameron Green, Josh Hazlewood, Usman Khawaja, Marnus Labuschagne, Nathan Lyon, Todd Murphy, Steven Smith (vice-capt), Mitchell Starc, David Warner.
Not sure if Scott Boland would fill in, instead of Todd Murphy 1
Let me give you a lay-of-the-land (post) below
The post below is organized into the following parts
- Analysis of Indian WTC batsmen from Jan 2016 – May 2023
- Analysis of Indian WTC batsmen against Australia from Jan 2016 -May 2023
- Analysis of Australian WTC batsmen from Jan 2016 – May 2023
- Analysis of Australian WTC batsmen against India from Jan 2016 -May 2023
- Analysis of Indian WTC bowlers from Jan 2016 – May 2023
- Analysis of Indian WTC bowlers against Australia from Jan 2016 -May 2023
- Analysis of Australian WTC bowlers from Jan 2016 – May 2023
- Analysis of Australian WTC bowlers gainst India from Jan 2016 -May 2023
- Team analysis of India and Australia
All the above analysis use data from ESPN Statsguru and use my R pakage cricketr
The data for the different players have been obtained using calls such as the ones below.
# Get Shubman Gill's batting data
#shubman <-getPlayerData(1070173,dir=".",file="shubman.csv",type="batting",homeOrAway=c(1,2), result=c(1,2,4))
#shubmansp <- getPlayerDataSp(1070173,tdir=".",tfile="shubmansp.csv",ttype="batting")
#Get Shubman Gill's data from Jan 2016 - May 2023
#df <-getPlayerDataHA(1070173,tfile="shubman1.csv",type="batting", matchType="Test")
#df1=getPlayerDataOppnHA(infile="shubman1.csv",outfile="shubmanTestAus.csv",startDate="2016-01-01",endDate="2023-05-01")
#Get Shubman Gills data from Jan 2016 - May 2023, against Australia
#df <-getPlayerDataHA(1070173,tfile="shubman1.csv",type="batting", matchType="Test")
#df1=getPlayerDataOppnHA(infile="shubman1.csv",outfile="shubmanTestAus.csv",opposition="Australia",startDate="2016-01-01",endDate="2023-05-01")
Note: To get data for bowlers we need to use the corresponding profile no and use type =‘bowling’. Details in my posts below
To do similar analysis please go through the following posts
- Re-introducing cricketr! : An R package to analyze performances of cricketers
- Cricketr learns new tricks : Performs fine-grained analysis of players
- Cricketr adds team analytics to its repertoire!!!
Note 1: I will not be analysing each and every chart as the charts are quite self-explanatory
Note 2: I have had to tile charts together otherwise this will become a very, very long post. You are free to use my R package cricketr and check out for yourself ##3. Analysis of India WTC batsmen from Jan 2016 – May 2023
Findings
- Kohli has the best average of 48+. India has won when Rohit and Rahane played well
- Kohli’s tops the list in cumulative average runs, followed by Pujara and Rohit is 3rd. Gill is on the upswing.
- Against Australia Pujara has the best cumulative average runs record followed by Rahane, with Gill in hot pursuit. In the strike rate department Gill tops followed by Rohit and Rahane
- Since 2016 Smith, Labuschagne has an average of 53+ since 2016!! Warner & Khwaja are at ~46
- Australia has won matches when Smith, Warner and Khwaja have played well.
- Labuschagne, Smith and C Green have good records against India. Indian bowlers will need to contain them
- Ashwin has the highest wickets followed by Jadeja against all teams. Ashwin’s performance has dropped over the years, while Siraj has been becoming better
- Jadeja has the best economy rate followed by Ashwin
- Against Australia specifically Jadeja has the best record followed by Ashwin. Jadeja has the best economy against Australia, followed by Siraj, then Ashwin
- Cummins, Starc and Lyons are the best performers for Australia. Hazzlewood, Cummins have the best economy against all opposition
- Against India Lyon, Cummins and Hazzlewood have performed well
- Hazzlewood, Lyon have a good economy rate against India
- Against Australia India has won 17 times, lost 60 and drawn 22 in Australia. At home India won 42, tied 2, lost 28 and drawn 24
- At the Oval where the World Test Championship is going to be held India has won 4, lost 10 and drawn 10.
Note 3: You can also read this post at Rpubs at ind-aus-WTC!! The formatting will be nicer!
Note 4: You can download this post as PDF to read at your leisure ind-aus-WTC.pdf
2. Install the cricketr package
if (!require("cricketr")){
install.packages("cricketr",lib = "c:/test")
}
library(cricketr)
3a. Basic analysis
The analyses below include – Runs frequency plot – Mean strike rate – Run Ranges
Kohli’s strike rate increases with increasing runs, while Gill’s seems to drop. So it is with Pujara & Rahane
par(mfrow=c(3,3))
par(mar=c(4,4,2,2))
batsmanRunsFreqPerf("kohliTest.csv","Kohli")
batsmanMeanStrikeRate("kohliTest.csv","Kohli")
batsmanRunsRanges("kohliTest.csv","Kohli")
batsmanRunsFreqPerf("rohitTest.csv","Rohit")
batsmanMeanStrikeRate("rohitTest.csv","Rohit")
batsmanRunsRanges("rohitTest.csv","Rohit")
batsmanRunsFreqPerf("shubmanTest.csv","S Gill")
batsmanMeanStrikeRate("shubmanTest.csv","S Gill")
batsmanRunsRanges("shubmanTest.csv","S Gill")

par(mfrow=c(2,3))
par(mar=c(4,4,2,2))
batsmanRunsFreqPerf("rahaneTest.csv","Rahane")
batsmanMeanStrikeRate("rahaneTest.csv","Rahane")
batsmanRunsRanges("rahaneTest.csv","Rahane")
batsmanRunsFreqPerf("pujaraTest.csv","Pujara")
batsmanMeanStrikeRate("pujaraTest.csv","Pujara")
batsmanRunsRanges("pujaraTest.csv","Pujara")

3b. More analyses
Kohli hits roughly 5 4s in his 50 versus Gill,Pujara who is able to smash 6 4s.
par(mfrow=c(3,3))
par(mar=c(4,4,2,2))
batsman4s("kohliTest.csv","Kohli")
batsman6s("kohliTest.csv","Kohli")
batsmanMeanStrikeRate("kohliTest.csv","Kohli")
batsman4s("rohitTest.csv","Rohit")
batsman6s("rohitTest.csv","Rohit")
batsmanMeanStrikeRate("rohitTest.csv","Rohit")
batsman4s("shubmanTest.csv","S Gill")
batsman6s("shubmanTest.csv","S Gill")
batsmanMeanStrikeRate("shubmanTest.csv","S Gill")

par(mfrow=c(2,3))
par(mar=c(4,4,2,2))
batsman4s("rahaneTest.csv","Rahane")
batsman6s("rahaneTest.csv","Rahane")
batsmanMeanStrikeRate("rahane.csv","Rahane")
batsman4s("pujaraTest.csv","Pujara")
batsman6s("pujaraTest.csv","Pujara")
batsmanMeanStrikeRate("pujaraTest.csv","Pujara")

3c.Boxplot histogram plot
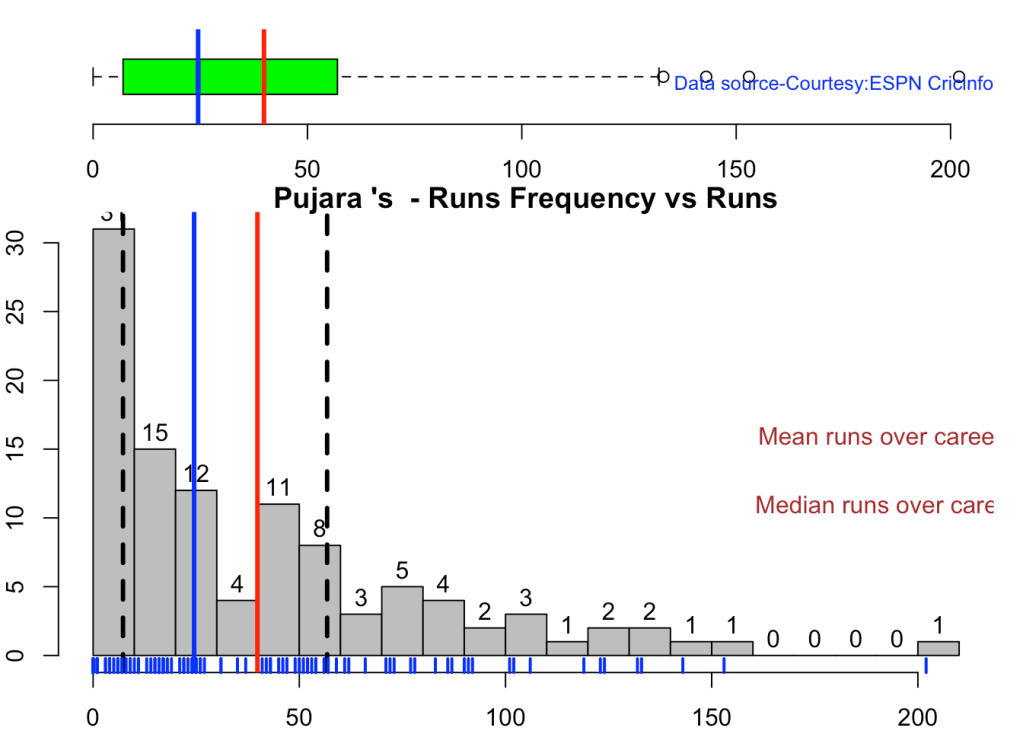
This plot shows a combined boxplot of the Runs ranges and a histog2ram of the Runs Frequency Kohli’s average is 48, while Rohit,Pujara is 40 with Rahane and Gill around 33.
batsmanPerfBoxHist("kohliTest.csv","Kohli")

batsmanPerfBoxHist("rohitTest.csv","Rohit")

batsmanPerfBoxHist("shubmanTest.csv","S Gill")
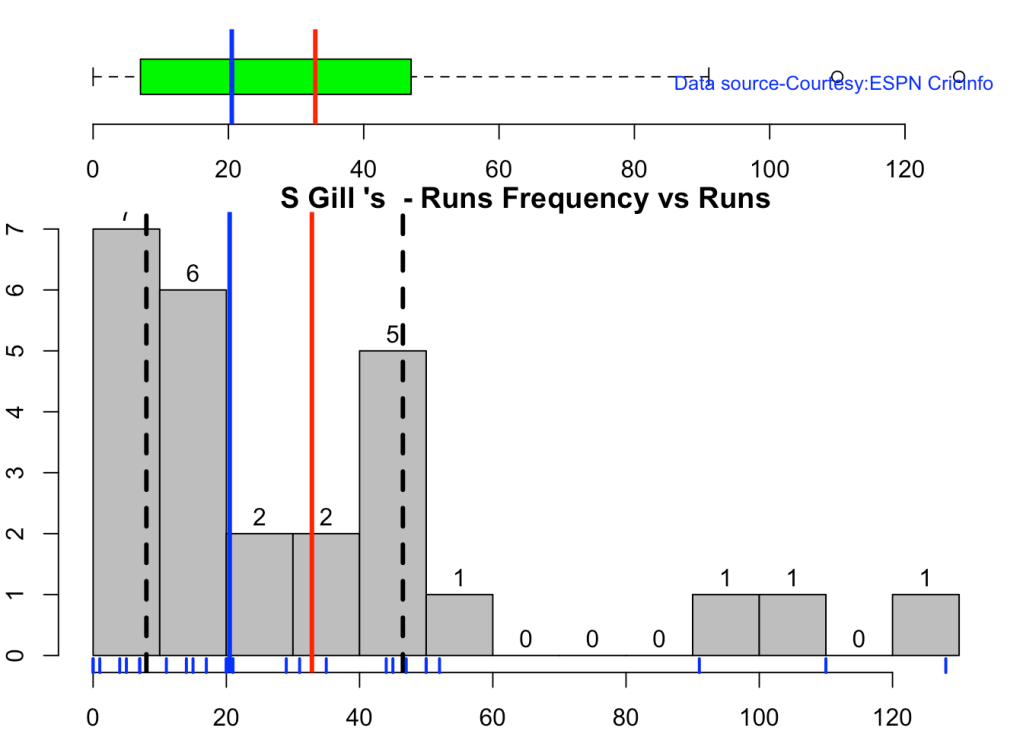
batsmanPerfBoxHist("rahaneTest.csv","Rahane")

batsmanPerfBoxHist("pujaraTest.csv","Pujara")
3d. Contribution to won and lost matches
For the functions below you will have to use the getPlayerDataSp() function. When Rohit Sharma and Pujara have played well India have tended to win more often
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
batsmanContributionWonLost("kohlisp.csv","Kohli")
batsmanContributionWonLost("rohitsp.csv","Rohit")
batsmanContributionWonLost("rahanesp.csv","Rahane")
batsmanContributionWonLost("pujarasp.csv","Pujara")

3e. Performance at home and overseas
This function also requires the use of getPlayerDataSp() as shown above. This can only be used for Test matches
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
batsmanPerfHomeAway("kohlisp.csv","Kohli")
batsmanPerfHomeAway("rohitsp.csv","Rohit")
batsmanPerfHomeAway("rahanesp.csv","Rahane")
batsmanPerfHomeAway("pujarasp.csv","Pujara")

3f. Batsman average at different venues
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
batsmanAvgRunsGround("kohliTest.csv","Kohli")
batsmanAvgRunsGround("rohitTest.csv","Rohit")
batsmanAvgRunsGround("rahaneTest.csv","Rahane")
batsmanAvgRunsGround("pujaraTest.csv","Pujara")
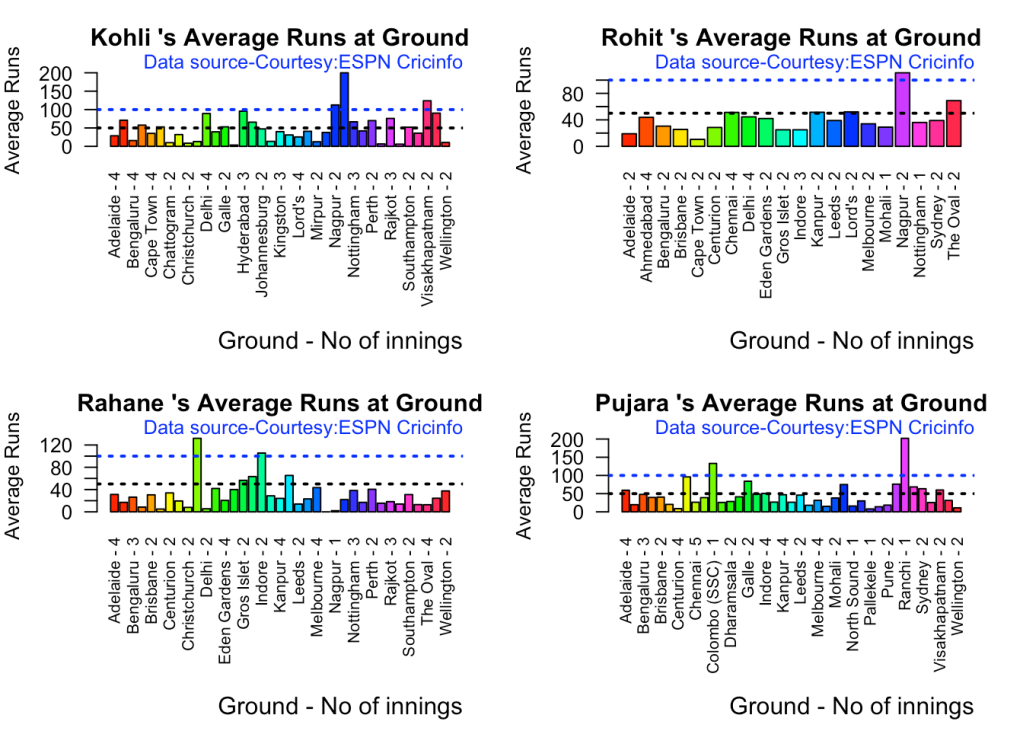
3g. Batsman average against different opposition
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
batsmanAvgRunsOpposition("kohliTest.csv","Kohli")
batsmanAvgRunsOpposition("rohitTest.csv","Rohit")
batsmanAvgRunsOpposition("rahaneTest.csv","Rahane")
batsmanAvgRunsOpposition("pujaraTest.csv","Pujara")
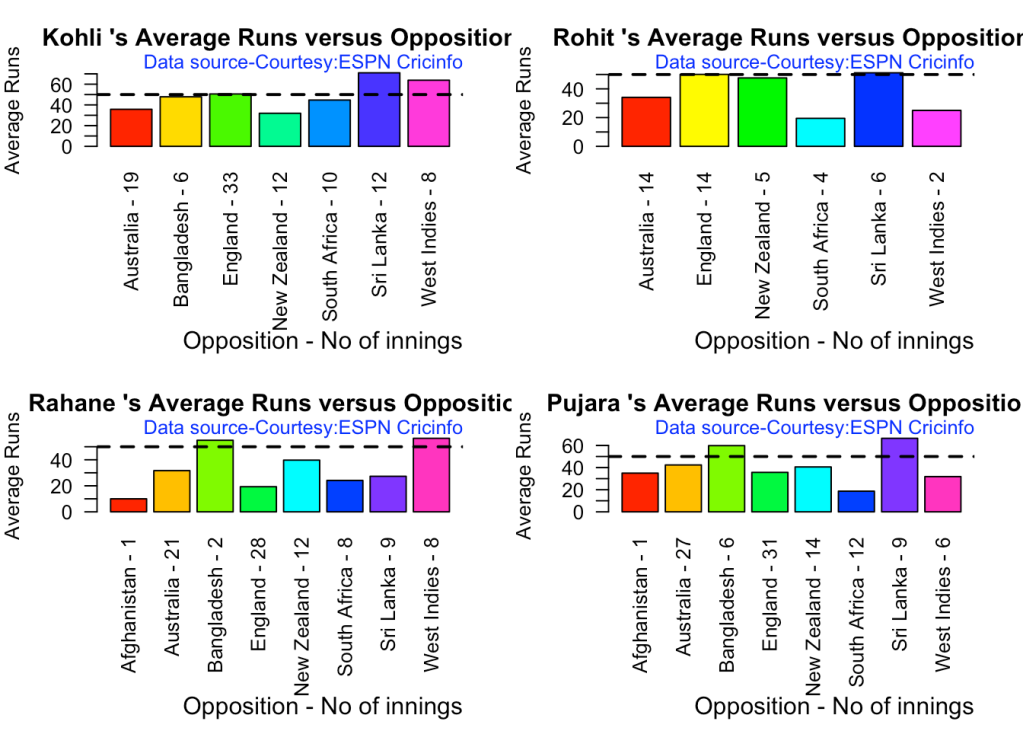
3h. Runs Likelihood of batsman
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
batsmanRunsLikelihood("kohli.csv","Kohli")
## Summary of Kohli 's runs scoring likelihood
## **************************************************
##
## There is a 52.91 % likelihood that Kohli will make 12 Runs in 26 balls over 35 Minutes
## There is a 30.81 % likelihood that Kohli will make 52 Runs in 100 balls over 139 Minutes
## There is a 16.28 % likelihood that Kohli will make 142 Runs in 237 balls over 335 Minutes
batsmanRunsLikelihood("rohit.csv","Rohit")
## Summary of Rohit 's runs scoring likelihood
## **************************************************
##
## There is a 43.24 % likelihood that Rohit will make 10 Runs in 21 balls over 32 Minutes
## There is a 45.95 % likelihood that Rohit will make 46 Runs in 85 balls over 124 Minutes
## There is a 10.81 % likelihood that Rohit will make 110 Runs in 199 balls over 282 Minutes
batsmanRunsLikelihood("rahane.csv","Rahane")
## Summary of Rahane 's runs scoring likelihood
## **************************************************
##
## There is a 7.75 % likelihood that Rahane will make 124 Runs in 224 balls over 318 Minutes
## There is a 62.02 % likelihood that Rahane will make 12 Runs in 26 balls over 37 Minutes
## There is a 30.23 % likelihood that Rahane will make 55 Runs in 113 balls over 162 Minutes
batsmanRunsLikelihood("pujara.csv","Pujara")
## Summary of Pujara 's runs scoring likelihood
## **************************************************
##
## There is a 60.49 % likelihood that Pujara will make 15 Runs in 38 balls over 55 Minutes
## There is a 31.48 % likelihood that Pujara will make 62 Runs in 142 balls over 204 Minutes
## There is a 8.02 % likelihood that Pujara will make 153 Runs in 319 balls over 445 Minutes

3h1. Moving average of batsman
Kohli’s moving average in tests seem to havw dropped after a peak in 2017, 2018. So it is with Rahane
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
batsmanMovingAverage("kohli.csv","Kohli")
batsmanMovingAverage("rohit.csv","Rohit")
batsmanMovingAverage("rahane.csv","Rahane")
batsmanMovingAverage("pujara.csv","Pujara")

3i. Cumulative Average runs of batsman in career
Kohli’s cumulative average averages to ~48. Shubman Gill’s cumulative average is on the rise.
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
batsmanCumulativeAverageRuns("kohliTest.csv","Kohli")

batsmanCumulativeAverageRuns("rohitTest.csv","Rohit")
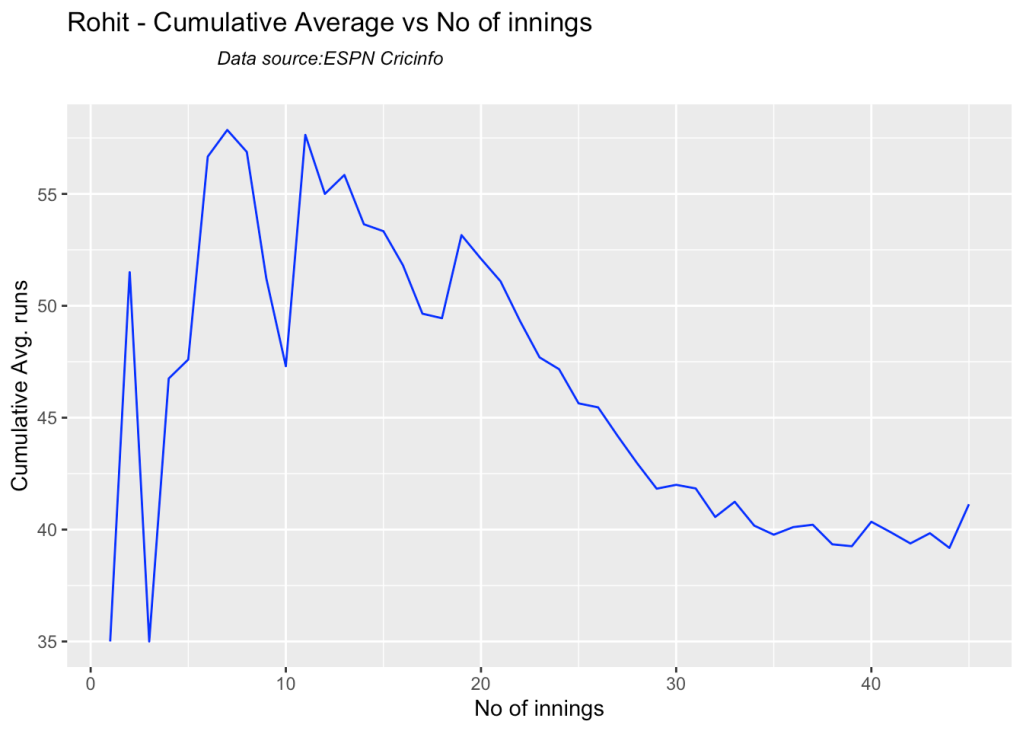
batsmanCumulativeAverageRuns("rahaneTest.csv","Rahane")
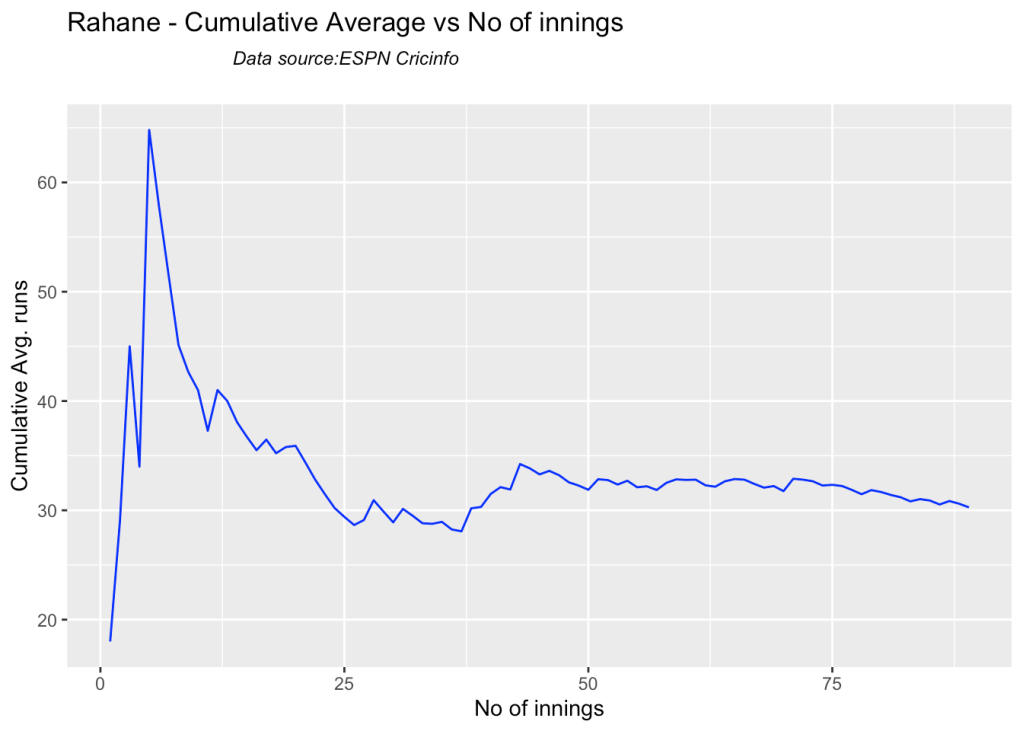
batsmanCumulativeAverageRuns("pujaraTest.csv","Pujara")

batsmanCumulativeAverageRuns("shubmanTest.csv","S Gill")

3j Cumulative Average strike rate of batsman in career
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
batsmanCumulativeStrikeRate("kohliTest.csv","Kohli")

batsmanCumulativeStrikeRate("rohitTest.csv","Rohit")

batsmanCumulativeStrikeRate("rahaneTest.csv","Rahane")

batsmanCumulativeStrikeRate("pujaraTest.csv","Pujara")

batsmanCumulativeStrikeRate("shubmanTest.csv","S Gill")

3k. Future Runs forecast
Here are plots that forecast how the batsman will perform in future. In this case 90% of the career runs trend is uses as the training set. the remaining 10% is the test set.
A Holt-Winters forecating model is used to forecast future performance based on the 90% training set. The forecated runs trend is plotted. The test set is also plotted to see how close the forecast and the actual matches
Take a look at the runs forecasted for the batsman below.
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
batsmanPerfForecast("kohli.csv","Kohli")
batsmanPerfForecast("rohit.csv","Rohit")
batsmanPerfForecast("rahane.csv","Rahane")
batsmanPerfForecast("pujara.csv","Pujara")

3l. Relative Mean Strike Rate plot
The plot below compares the Mean Strike Rate of the batsman for each of the runs ranges of 10 and plots them. The plot indicate the following
frames <- list("kohliTest.csv","rohitTest.csv","pujaraTest.csv","rahaneTest.csv","shubmanTest.csv")
names <- list("Kohli","Rohit","Pujara","Rahane","S Gill")
relativeBatsmanSR(frames,names)

3m. Relative Runs Frequency plot
The plot below gives the relative Runs Frequency Percetages for each 10 run bucket. The plot below show
frames <- list("kohliTest.csv","rohitTest.csv","pujaraTest.csv","rahaneTest.csv","shubmanTest.csv")
names <- list("Kohli","Rohit","Pujara","Rahane","S Gill")
relativeRunsFreqPerf(frames,names)

3n. Relative cumulative average runs in career
Kohli’s tops the list, followed by Pujara and Rohit is 3rd. Gill is on the upswing. Hope he performs well.
frames <- list("kohliTest.csv","rohitTest.csv","pujaraTest.csv","rahaneTest.csv","shubmanTest.csv")
names <- list("Kohli","Rohit","Pujara","Rahane","S Gill")
relativeBatsmanCumulativeAvgRuns(frames,names)

3o. Relative cumulative average strike rate in career
ROhit has the best strike rate followed by Kohli, with Shubman Gill ctaching up fast
frames <- list("kohliTest.csv","rohitTest.csv","pujaraTest.csv","rahaneTest.csv","shubmanTest.csv")
names <- list("Kohli","Rohit","Pujara","Rahane","S Gill")
relativeBatsmanCumulativeStrikeRate(frames,names)

3p. Check Batsman In-Form or Out-of-Form
The below computation uses Null Hypothesis testing and p-value to determine if the batsman is in-form or out-of-form. For this 90% of the career runs is chosen as the population and the mean computed. The last 10% is chosen to be the sample set and the sample Mean and the sample Standard Deviation are caculated.
The Null Hypothesis (H0) assumes that the batsman continues to stay in-form where the sample mean is within 95% confidence interval of population mean The Alternative (Ha) assumes that the batsman is out of form the sample mean is beyond the 95% confidence interval of the population mean.
A significance value of 0.05 is chosen and p-value us computed If p-value >= .05 – Batsman In-Form If p-value < 0.05 – Batsman Out-of-Form
Note Ideally the p-value should be done for a population that follows the Normal Distribution. But the runs population is usually left skewed. So some correction may be needed. I will revisit this later
This is done for the Top 4 batsman
checkBatsmanInForm("kohli.csv","Kohli")
## [1] "**************************** Form status of Kohli ****************************\n\n Population size: 154 Mean of population: 47.03 \n Sample size: 18 Mean of sample: 32.22 SD of sample: 42.45 \n\n Null hypothesis H0 : Kohli 's sample average is within 95% confidence interval of population average\n Alternative hypothesis Ha : Kohli 's sample average is below the 95% confidence interval of population average\n\n Kohli 's Form Status: In-Form because the p value: 0.078058 is greater than alpha= 0.05 \n *******************************************************************************************\n\n"
checkBatsmanInForm("rohit.csv","Rohit")
## [1] "**************************** Form status of Rohit ****************************\n\n Population size: 66 Mean of population: 37.03 \n Sample size: 8 Mean of sample: 37.88 SD of sample: 35.38 \n\n Null hypothesis H0 : Rohit 's sample average is within 95% confidence interval of population average\n Alternative hypothesis Ha : Rohit 's sample average is below the 95% confidence interval of population average\n\n Rohit 's Form Status: In-Form because the p value: 0.526254 is greater than alpha= 0.05 \n *******************************************************************************************\n\n"
checkBatsmanInForm("rahane.csv","Rahane")
## [1] "**************************** Form status of Rahane ****************************\n\n Population size: 116 Mean of population: 34.78 \n Sample size: 13 Mean of sample: 21.38 SD of sample: 21.96 \n\n Null hypothesis H0 : Rahane 's sample average is within 95% confidence interval of population average\n Alternative hypothesis Ha : Rahane 's sample average is below the 95% confidence interval of population average\n\n Rahane 's Form Status: Out-of-Form because the p value: 0.023244 is less than alpha= 0.05 \n *******************************************************************************************\n\n"
checkBatsmanInForm("pujara.csv","Pujara")
## [1] "**************************** Form status of Pujara ****************************\n\n Population size: 145 Mean of population: 41.93 \n Sample size: 17 Mean of sample: 33.24 SD of sample: 31.74 \n\n Null hypothesis H0 : Pujara 's sample average is within 95% confidence interval of population average\n Alternative hypothesis Ha : Pujara 's sample average is below the 95% confidence interval of population average\n\n Pujara 's Form Status: In-Form because the p value: 0.137319 is greater than alpha= 0.05 \n *******************************************************************************************\n\n"
checkBatsmanInForm("shubman.csv","S Gill")
## [1] "**************************** Form status of S Gill ****************************\n\n Population size: 23 Mean of population: 30.43 \n Sample size: 3 Mean of sample: 51.33 SD of sample: 66.88 \n\n Null hypothesis H0 : S Gill 's sample average is within 95% confidence interval of population average\n Alternative hypothesis Ha : S Gill 's sample average is below the 95% confidence interval of population average\n\n S Gill 's Form Status: In-Form because the p value: 0.687033 is greater than alpha= 0.05 \n *******************************************************************************************\n\n"
3q. Predicting Runs given Balls Faced and Minutes at Crease
A multi-variate regression plane is fitted between Runs and Balls faced +Minutes at crease.
BF <- seq( 10, 400,length=15)
Mins <- seq(30,600,length=15)
newDF <- data.frame(BF,Mins)
kohli1 <- batsmanRunsPredict("kohli.csv","Kohli",newdataframe=newDF)
rohit1 <- batsmanRunsPredict("rohit.csv","Rohit",newdataframe=newDF)
pujara1 <- batsmanRunsPredict("pujara.csv","Pujara",newdataframe=newDF)
rahane1 <- batsmanRunsPredict("rahane.csv","Rahane",newdataframe=newDF)
sgill1 <- batsmanRunsPredict("shubman.csv","S Gill",newdataframe=newDF)
batsmen <-cbind(round(kohli1$Runs),round(rohit1$Runs),round(pujara1$Runs),round(rahane1$Runs),round(sgill1$Runs))
colnames(batsmen) <- c("Kohli","Rohit","Pujara","Rahane","S Gill")
newDF <- data.frame(round(newDF$BF),round(newDF$Mins))
colnames(newDF) <- c("BallsFaced","MinsAtCrease")
predictedRuns <- cbind(newDF,batsmen)
predictedRuns
## BallsFaced MinsAtCrease Kohli Rohit Pujara Rahane S Gill
## 1 10 30 6 3 3 2 7
## 2 38 71 24 19 16 17 24
## 3 66 111 41 35 29 31 40
## 4 94 152 58 51 42 45 56
## 5 121 193 76 66 55 59 73
## 6 149 234 93 82 68 74 89
## 7 177 274 110 98 80 88 106
## 8 205 315 128 114 93 102 122
## 9 233 356 145 129 106 116 139
## 10 261 396 163 145 119 130 155
## 11 289 437 180 161 132 145 171
## 12 316 478 197 177 144 159 188
## 13 344 519 215 192 157 173 204
## 14 372 559 232 208 170 187 221
## 15 400 600 249 224 183 202 237
4. Analysis of India WTC batsmen from Jan 2016 – May 2023 against Australia
4a. Relative cumulative average
Against Australia specifically between 2016 – 2023, Pujara has the best record followed by Rahane, with Gill in hot pursuit. Kohli and Rohit trail behind
frames <- list("kohliTestAus.csv","rohitTestAus.csv","pujaraTestAus.csv","rahaneTestAus.csv","shubmanTestAus.csv")
names <- list("Kohli","Rohit","Pujara","Rahane","S Gill")
relativeBatsmanCumulativeAvgRuns(frames,names)

4b. Relative cumulative average strike rate in career
In the Strike Rate department Gill tops followed by Rohit and Rahane
frames <- list("kohliTestAus.csv","rohitTestAus.csv","pujaraTestAus.csv","rahaneTestAus.csv","shubmanTestAus.csv")
names <- list("Kohli","Rohit","Pujara","Rahane","S Gill")
relativeBatsmanCumulativeStrikeRate(frames,names)

5. Analysis of Australia WTC batsmen from Jan 2016 – May 2023
5a Basic analyses
par(mfrow=c(3,3))
par(mar=c(4,4,2,2))
batsmanRunsFreqPerf("stevesmithTest.csv","S Smith")
batsmanMeanStrikeRate("stevesmithTest.csv","S Smith")
batsmanRunsRanges("stevesmithTest.csv","S Smith")
batsmanRunsFreqPerf("warnerTest.csv","Warner")
batsmanMeanStrikeRate("warnerTest.csv","Warner")
batsmanRunsRanges("warnerTest.csv","Warner")
batsmanRunsFreqPerf("labuschagneTest.csv","M Labuschagne")
batsmanMeanStrikeRate("labuschagneTest.csv","M Labuschagne")
batsmanRunsRanges("labuschagneTest.csv","M Labuschagne")

par(mfrow=c(2,3))
par(mar=c(4,4,2,2))
batsmanRunsFreqPerf("cgreenTest.csv","C Green")
batsmanMeanStrikeRate("cgreenTest.csv","C Green")
batsmanRunsRanges("cgreenTest.csv","C Green")
batsmanRunsFreqPerf("khwajaTest.csv","Khwaja")
batsmanMeanStrikeRate("khwajaTest.csv","Khwaja")
batsmanRunsRanges("khwajaTest.csv","Khwaja")

5b. More analyses
par(mfrow=c(3,3))
par(mar=c(4,4,2,2))
batsman4s("stevesmithTest.csv","S Smith")
batsman6s("stevesmithTest.csv","S Smith")
batsmanMeanStrikeRate("stevesmithTest.csv","S Smith")
batsman4s("warnerTest.csv","Warner")
batsman6s("warnerTest.csv","Warner")
batsmanMeanStrikeRate("warnerTest.csv","Warner")
batsman4s("labuschagneTest.csv","M Labuschagne")
batsman6s("labuschagneTest.csv","M Labuschagne")
batsmanMeanStrikeRate("labuschagneTest.csv","M Labuschagne")

par(mfrow=c(2,3))
par(mar=c(4,4,2,2))
batsman4s("cgreenTest.csv","C Green")
batsman6s("cgreenTest.csv","C Green")
batsmanMeanStrikeRate("cgreenTest.csv","C Green")
batsman4s("khwajaTest.csv","Khwaja")
batsman6s("khwajaTest.csv","Khwaja")
batsmanMeanStrikeRate("khwajaTest.csv","Khwaja")

5c.Boxplot histogram plot
This plot shows a combined boxplot of the Runs ranges and a histog2ram of the Runs Frequency
Smith, Labuschagne has an average of 53+ since 2016!! Warner & Khwaja are at ~46
batsmanPerfBoxHist("stevesmithTest.csv","S Smith")

batsmanPerfBoxHist("warnerTest.csv","Warner")
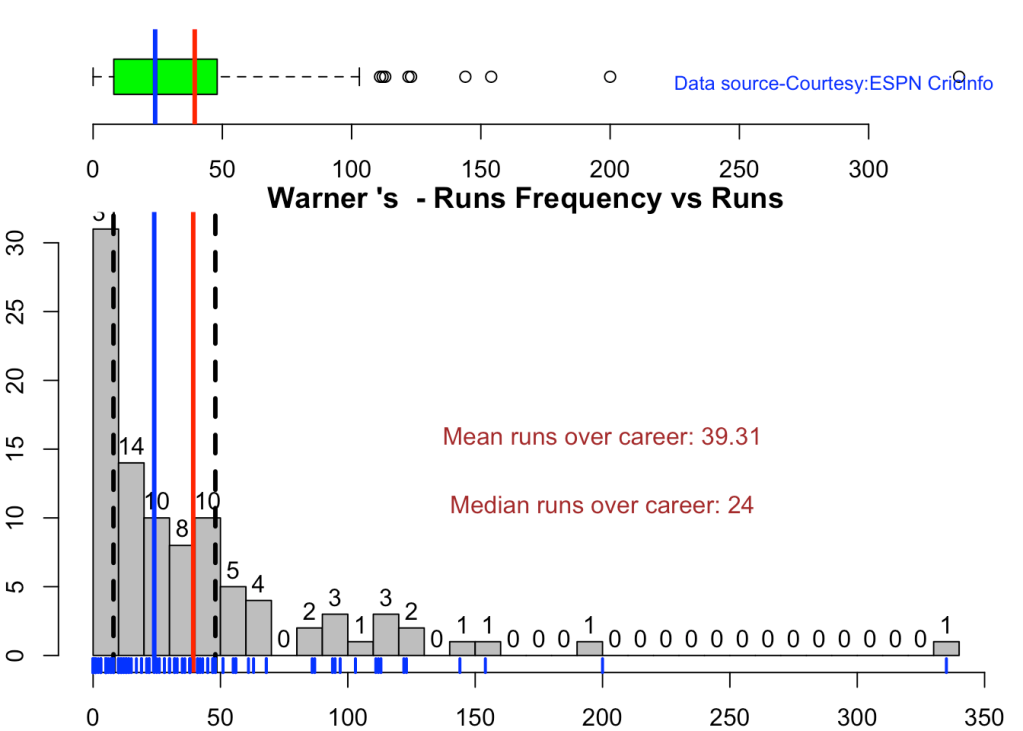
batsmanPerfBoxHist("labuschagneTest.csv","M Labuschagne")

batsmanPerfBoxHist("cgreenTest.csv","C Green")

batsmanPerfBoxHist("khwajaTest.csv","Khwaja")
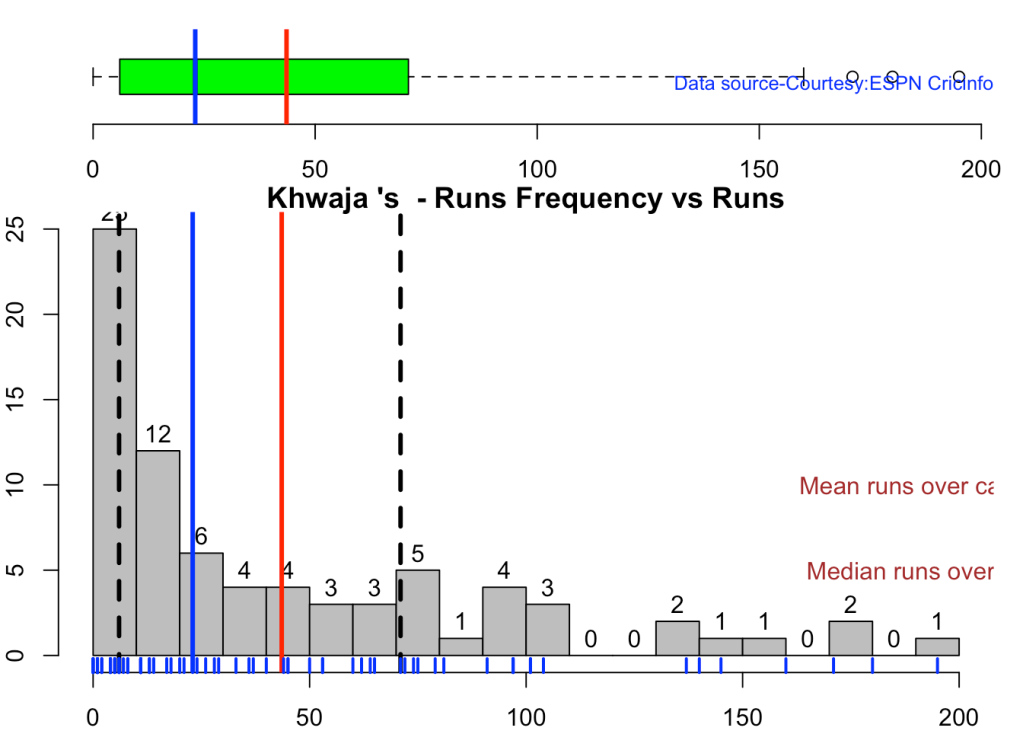
5d. Contribution to won and lost matches
For the 2 functions below you will have to use the getPlayerDataSp() function. Australia has won matches when Smith, Warner and Khwaja have played well.
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
batsmanContributionWonLost("stevesmithsp.csv","S Smith")
batsmanContributionWonLost("warnersp.csv","Warner")
batsmanContributionWonLost("labuschagnesp.csv","M Labuschagne")
batsmanContributionWonLost("cgreensp.csv","C Green")

batsmanContributionWonLost("khwajasp.csv","Khwaja")

5e. Performance at home and overseas
This function also requires the use of getPlayerDataSp() as shown above. This can only be used for Test matches
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
batsmanPerfHomeAway("stevesmithsp.csv","S Smith")
batsmanPerfHomeAway("warnersp.csv","Warner")
batsmanPerfHomeAway("labuschagnesp.csv","M Labuschagne")
batsmanPerfHomeAway("cgreensp.csv","C Green")

batsmanPerfHomeAway("khwajasp.csv","Khwaja")

5f. Batsman average at different venues
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
batsmanAvgRunsGround("stevesmithTest.csv","S Smith")
batsmanAvgRunsGround("warnerTest.csv","Warner")
batsmanAvgRunsGround("labuschagneTest.csv","M Labuschagne")
batsmanAvgRunsGround("cgreenTest.csv","C Green")

batsmanAvgRunsGround("khwajaTest.csv","Khwaja")

5g. Batsman average against different opposition
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
batsmanAvgRunsOpposition("stevesmithTest.csv","S Smith")
batsmanAvgRunsOpposition("warnerTest.csv","Warner")
batsmanAvgRunsOpposition("labuschagneTest.csv","M Labuschagne")
batsmanAvgRunsOpposition("khwajaTest.csv","Khwaja")

5h. Runs Likelihood of batsman
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
batsmanRunsLikelihood("stevesmithTest.csv","S Smith")
## Summary of S Smith 's runs scoring likelihood
## **************************************************
##
## There is a 58.76 % likelihood that S Smith will make 21 Runs in 38 balls over 56 Minutes
## There is a 24.74 % likelihood that S Smith will make 70 Runs in 148 balls over 210 Minutes
## There is a 16.49 % likelihood that S Smith will make 148 Runs in 268 balls over 398 Minutes
batsmanRunsLikelihood("warnerTest.csv","Warner")
## Summary of Warner 's runs scoring likelihood
## **************************************************
##
## There is a 7.22 % likelihood that Warner will make 155 Runs in 253 balls over 372 Minutes
## There is a 62.89 % likelihood that Warner will make 14 Runs in 21 balls over 32 Minutes
## There is a 29.9 % likelihood that Warner will make 65 Runs in 94 balls over 135 Minutes
batsmanRunsLikelihood("labuschagneTest.csv","M Labuschagne")
## Summary of M Labuschagne 's runs scoring likelihood
## **************************************************
##
## There is a 32.76 % likelihood that M Labuschagne will make 74 Runs in 144 balls over 206 Minutes
## There is a 55.17 % likelihood that M Labuschagne will make 22 Runs in 37 balls over 54 Minutes
## There is a 12.07 % likelihood that M Labuschagne will make 168 Runs in 297 balls over 420 Minutes

batsmanRunsLikelihood("khwajaTest.csv","Khwaja")
## Summary of Khwaja 's runs scoring likelihood
## **************************************************
##
## There is a 64.94 % likelihood that Khwaja will make 14 Runs in 29 balls over 42 Minutes
## There is a 27.27 % likelihood that Khwaja will make 79 Runs in 148 balls over 210 Minutes
## There is a 7.79 % likelihood that Khwaja will make 165 Runs in 351 balls over 515 Minutes
5i. Moving average of batsman
Smith and Warner’s moving average has been on a downward trend lately. Khwaja is playing well
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
batsmanMovingAverage("stevesmith.csv","S Smith")
batsmanMovingAverage("warner.csv","Warner")
batsmanMovingAverage("labuschagne.csv","M Labuschagne")
batsmanMovingAverage("khwaja.csv","Khwaja")

5j. Cumulative Average runs of batsman in career
Labuschagne, SMith and Warner havwe very good cumulative average
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
batsmanCumulativeAverageRuns("stevesmithTest.csv","S Smith")

batsmanCumulativeAverageRuns("warnerTest.csv","Warner")
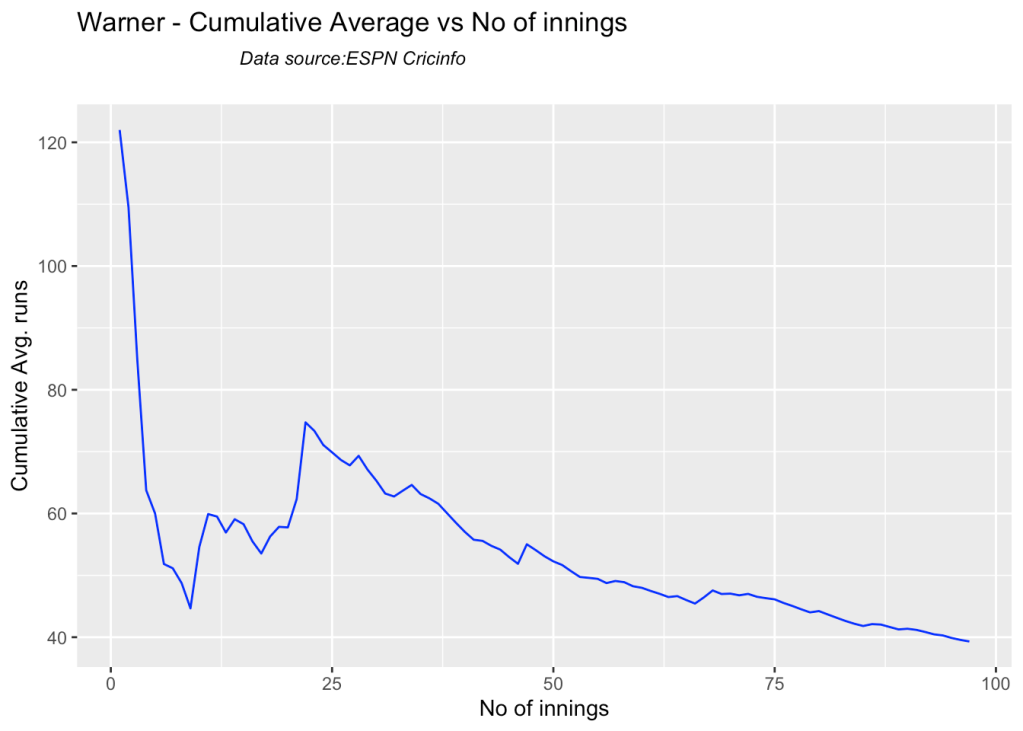
batsmanCumulativeAverageRuns("labuschagneTest.csv","M Labuschagne")

batsmanCumulativeAverageRuns("khwajaTest.csv","Khwaja")

5k. Cumulative Average strike rate of batsman in career
Warner towers over the others in the cumulative strike rate, followed by Labuschagne and Smith
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
batsmanCumulativeStrikeRate("stevesmithTest.csv","S Smith")

batsmanCumulativeStrikeRate("warnerTest.csv","Warner")
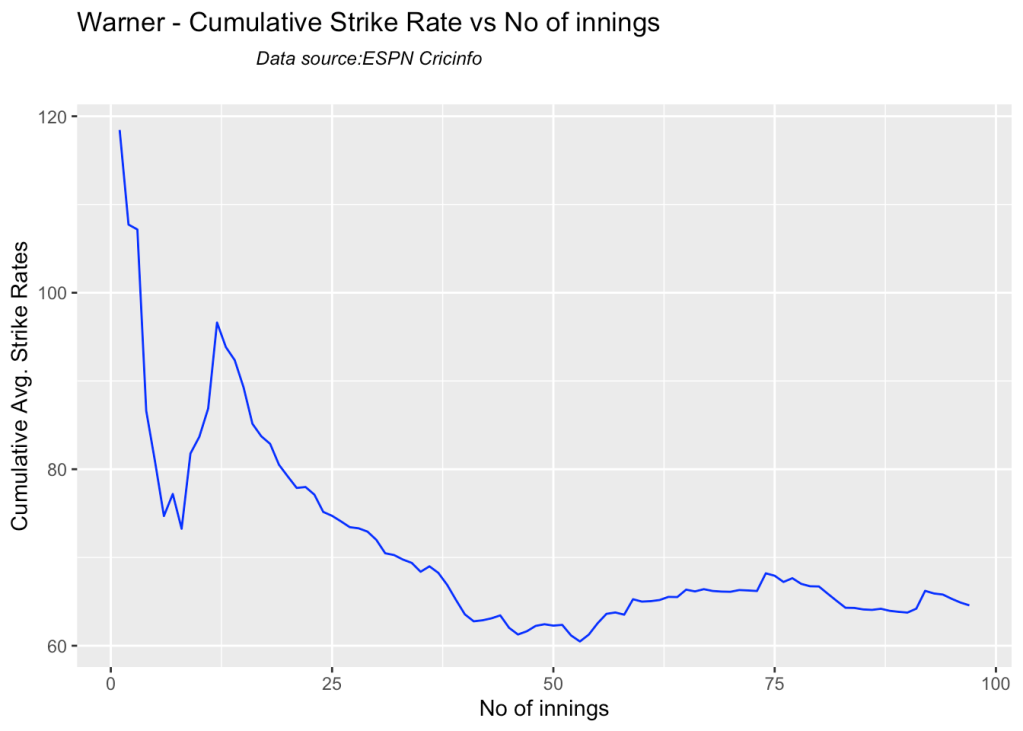
batsmanCumulativeStrikeRate("labuschagneTest.csv","M Labuschagne")

batsmanCumulativeStrikeRate("khwajaTest.csv","Khwaja")

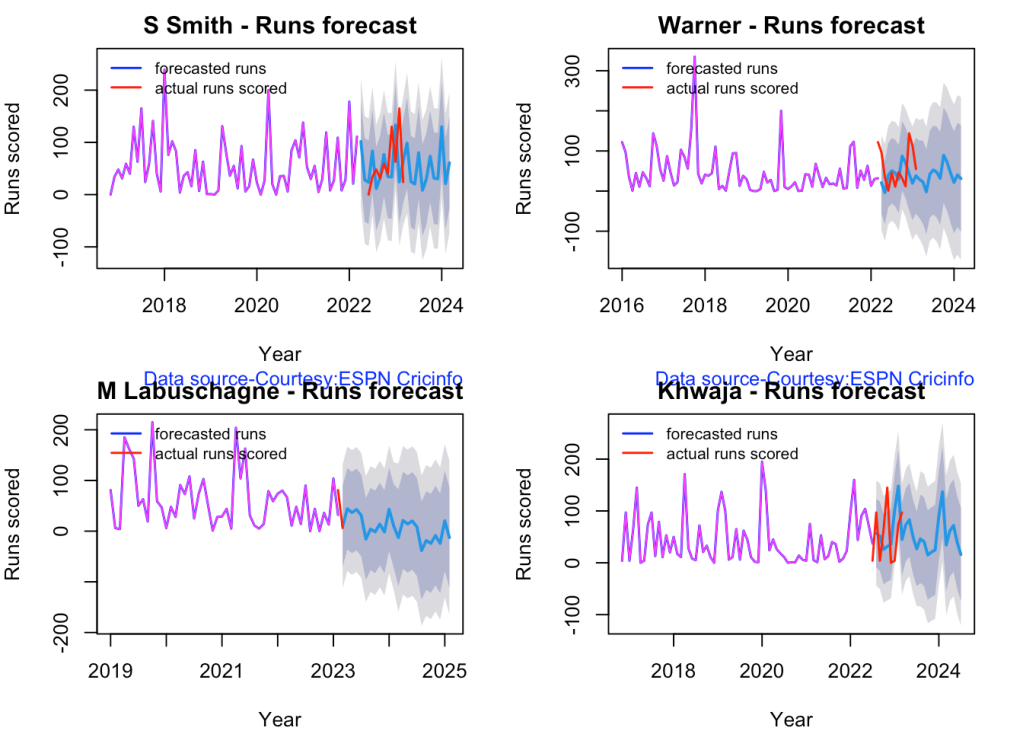
5l. Future Runs forecast
Here are plots that forecast how the batsman will perform in future. In this case 90% of the career runs trend is uses as the training set. the remaining 10% is the test set.
A Holt-Winters forecating model is used to forecast future performance based on the 90% training set. The forecated runs trend is plotted. The test set is also plotted to see how close the forecast and the actual matches
Take a look at the runs forecasted for the batsman below.
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
batsmanPerfForecast("stevesmithTest.csv","S Smith")
batsmanPerfForecast("warnerTest.csv","Warner")
batsmanPerfForecast("labuschagneTest.csv","M Labuschagne")
batsmanPerfForecast("khwajaTest.csv","Khwaja")
5m. Relative Mean Strike Rate plot
The plot below compares the Mean Strike Rate of the batsman for each of the runs ranges of 10 and plots them. The plot indicate the following
frames <- list("stevesmithTest.csv","warnerTest.csv","khwajaTest.csv","labuschagneTest.csv","cgreenTest.csv")
names <- list("S Smith","Warner","Khwaja","Labuschagne","C Green")
relativeBatsmanSR(frames,names)

5n. Relative Runs Frequency plot
The plot below gives the relative Runs Frequency Percetages for each 10 run bucket. The plot below show
frames <- list("stevesmithTest.csv","warnerTest.csv","khwajaTest.csv","labuschagneTest.csv","cgreenTest.csv")
names <- list("S Smith","Warner","Khwaja","Labuschagne","C Green")
relativeRunsFreqPerf(frames,names)

5o. Relative cumulative average runs in career
frames <- list("stevesmithTest.csv","warnerTest.csv","khwajaTest.csv","labuschagneTest.csv","cgreenTest.csv")
names <- list("S Smith","Warner","Khwaja","Labuschagne","C Green")
relativeBatsmanCumulativeAvgRuns(frames,names)

5p. Relative cumulative average strike rate in career
frames <- list("stevesmithTest.csv","warnerTest.csv","khwajaTest.csv","labuschagneTest.csv","cgreenTest.csv")
names <- list("S Smith","Warner","Khwaja","Labuschagne","C Green")
relativeBatsmanCumulativeStrikeRate(frames,names)

5q. Check Batsman In-Form or Out-of-Form
The below computation uses Null Hypothesis testing and p-value to determine if the batsman is in-form or out-of-form. For this 90% of the career runs is chosen as the population and the mean computed. The last 10% is chosen to be the sample set and the sample Mean and the sample Standard Deviation are caculated.
The Null Hypothesis (H0) assumes that the batsman continues to stay in-form where the sample mean is within 95% confidence interval of population mean The Alternative (Ha) assumes that the batsman is out of form the sample mean is beyond the 95% confidence interval of the population mean.
A significance value of 0.05 is chosen and p-value us computed If p-value >= .05 – Batsman In-Form If p-value < 0.05 – Batsman Out-of-Form
Note Ideally the p-value should be done for a population that follows the Normal Distribution. But the runs population is usually left skewed. So some correction may be needed. I will revisit this later
This is done for the Top 4 batsman
checkBatsmanInForm("stevesmith.csv","S Smith")
## [1] "**************************** Form status of S Smith ****************************\n\n Population size: 144 Mean of population: 53.76 \n Sample size: 17 Mean of sample: 45.65 SD of sample: 56.4 \n\n Null hypothesis H0 : S Smith 's sample average is within 95% confidence interval of population average\n Alternative hypothesis Ha : S Smith 's sample average is below the 95% confidence interval of population average\n\n S Smith 's Form Status: In-Form because the p value: 0.280533 is greater than alpha= 0.05 \n *******************************************************************************************\n\n"
checkBatsmanInForm("warner.csv","Warner")
## [1] "**************************** Form status of Warner ****************************\n\n Population size: 164 Mean of population: 45.2 \n Sample size: 19 Mean of sample: 26.63 SD of sample: 44.62 \n\n Null hypothesis H0 : Warner 's sample average is within 95% confidence interval of population average\n Alternative hypothesis Ha : Warner 's sample average is below the 95% confidence interval of population average\n\n Warner 's Form Status: Out-of-Form because the p value: 0.042744 is less than alpha= 0.05 \n *******************************************************************************************\n\n"
checkBatsmanInForm("labuschagne.csv","M Labuschagne")
## [1] "**************************** Form status of M Labuschagne ****************************\n\n Population size: 52 Mean of population: 59.56 \n Sample size: 6 Mean of sample: 29.67 SD of sample: 19.96 \n\n Null hypothesis H0 : M Labuschagne 's sample average is within 95% confidence interval of population average\n Alternative hypothesis Ha : M Labuschagne 's sample average is below the 95% confidence interval of population average\n\n M Labuschagne 's Form Status: Out-of-Form because the p value: 0.005239 is less than alpha= 0.05 \n *******************************************************************************************\n\n"
checkBatsmanInForm("khwaja.csv","Khwaja")
## [1] "**************************** Form status of Khwaja ****************************\n\n Population size: 89 Mean of population: 41.62 \n Sample size: 10 Mean of sample: 53.1 SD of sample: 76.34 \n\n Null hypothesis H0 : Khwaja 's sample average is within 95% confidence interval of population average\n Alternative hypothesis Ha : Khwaja 's sample average is below the 95% confidence interval of population average\n\n Khwaja 's Form Status: In-Form because the p value: 0.677691 is greater than alpha= 0.05 \n *******************************************************************************************\n\n"
5r. Predicting Runs given Balls Faced and Minutes at Crease
A multi-variate regression plane is fitted between Runs and Balls faced +Minutes at crease.
BF <- seq( 10, 400,length=15)
Mins <- seq(30,600,length=15)
newDF <- data.frame(BF,Mins)
ssmith1 <- batsmanRunsPredict("stevesmith.csv","S Smith",newdataframe=newDF)
warner1 <- batsmanRunsPredict("warner.csv","Warner",newdataframe=newDF)
khwaja1 <- batsmanRunsPredict("khwaja.csv","Khwaja",newdataframe=newDF)
labuschagne1 <- batsmanRunsPredict("labuschagne.csv","Labuschagne",newdataframe=newDF)
cgreen1 <- batsmanRunsPredict("cgreen.csv","C Green",newdataframe=newDF)
batsmen <-cbind(round(ssmith1$Runs),round(warner1$Runs),round(khwaja1$Runs),round(labuschagne1$Runs),round(cgreen1$Runs))
colnames(batsmen) <- c("S Smith","Warner","Khwaja","Labuschagne","C Green")
newDF <- data.frame(round(newDF$BF),round(newDF$Mins))
colnames(newDF) <- c("BallsFaced","MinsAtCrease")
predictedRuns <- cbind(newDF,batsmen)
predictedRuns
## BallsFaced MinsAtCrease S Smith Warner Khwaja Labuschagne C Green
## 1 10 30 7 10 10 9 13
## 2 38 71 23 30 24 24 29
## 3 66 111 38 50 38 40 44
## 4 94 152 53 70 53 55 60
## 5 121 193 69 90 67 70 75
## 6 149 234 84 110 81 85 91
## 7 177 274 100 130 95 100 106
## 8 205 315 115 150 109 116 122
## 9 233 356 130 170 123 131 137
## 10 261 396 146 190 137 146 153
## 11 289 437 161 210 151 161 168
## 12 316 478 177 230 165 176 184
## 13 344 519 192 250 179 192 199
## 14 372 559 207 270 193 207 215
## 15 400 600 223 290 207 222 230
6. Analysis of Australia WTC batsmen from Jan 2016 – May 2023 against India
6a. Relative cumulative average runs in career
Labuschagne, Smith and C Green have good records against India
frames <- list("stevesmithTestInd.csv","warnerTestInd.csv","khwajaTestInd.csv","labuschagneTestInd.csv","cgreenTestInd.csv")
names <- list("S Smith","Warner","Khwaja","Labuschagne","C Green")
relativeBatsmanCumulativeAvgRuns(frames,names)

6b. Relative cumulative average strike rate in career
Warner, Labuschagne and Smith have a good strike rate against India
frames <- list("stevesmithTestInd.csv","warnerTestInd.csv","khwajaTestInd.csv","labuschagneTestInd.csv","cgreenTestInd.csv")
names <- list("S Smith","Warner","Khwaja","Labuschagne","C Green")
relativeBatsmanCumulativeStrikeRate(frames,names)

7. Analysis of India WTC bowlers from Jan 2016 – May 2023
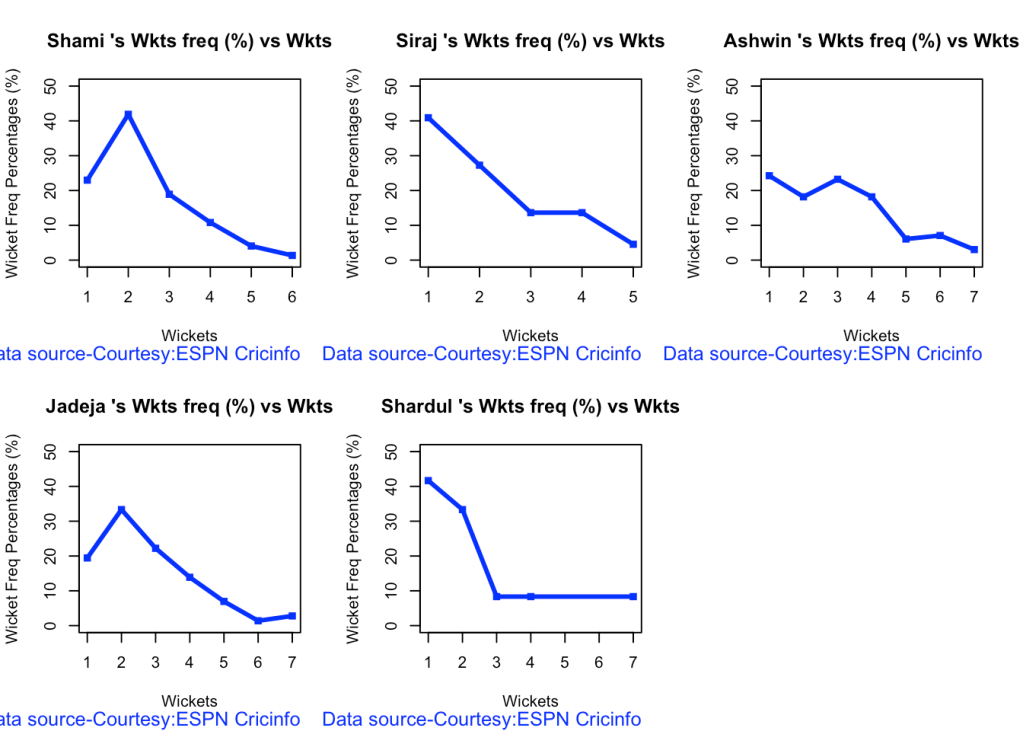
7a Wickets frequency chart
par(mfrow=c(2,3))
par(mar=c(4,4,2,2))
bowlerWktsFreqPercent("shamiTest.csv","Shami")
bowlerWktsFreqPercent("sirajTest.csv","Siraj")
bowlerWktsFreqPercent("ashwinTest.csv","Ashwin")
bowlerWktsFreqPercent("jadejaTest.csv","Jadeja")
bowlerWktsFreqPercent("shardulTest.csv","Shardul")
7b Wickets Runs chart
par(mfrow=c(2,3))
par(mar=c(4,4,2,2))
bowlerWktsRunsPlot("shamiTest.csv","Shami")
bowlerWktsRunsPlot("sirajTest.csv","Siraj")
bowlerWktsRunsPlot("ashwinTest.csv","Ashwin")
bowlerWktsRunsPlot("jadejaTest.csv","Jadeja")
bowlerWktsRunsPlot("shardulTest.csv","Shardul")

7c. Average wickets at different venues
par(mfrow=c(2,3))
par(mar=c(4,4,2,2))
bowlerAvgWktsGround("shamiTest.csv","Shami")
bowlerAvgWktsGround("sirajTest.csv","Siraj")
bowlerAvgWktsGround("ashwinTest.csv","Ashwin")
bowlerAvgWktsGround("jadejaTest.csv","Jadeja")
bowlerAvgWktsGround("shardulTest.csv","Shardul")

7d Average wickets against different opposition
par(mfrow=c(2,3))
par(mar=c(4,4,2,2))
bowlerAvgWktsOpposition("shamiTest.csv","Shami")
bowlerAvgWktsOpposition("sirajTest.csv","Siraj")
bowlerAvgWktsOpposition("ashwinTest.csv","Ashwin")
bowlerAvgWktsOpposition("jadejaTest.csv","Jadeja")
bowlerAvgWktsOpposition("shardulTest.csv","Shardul")
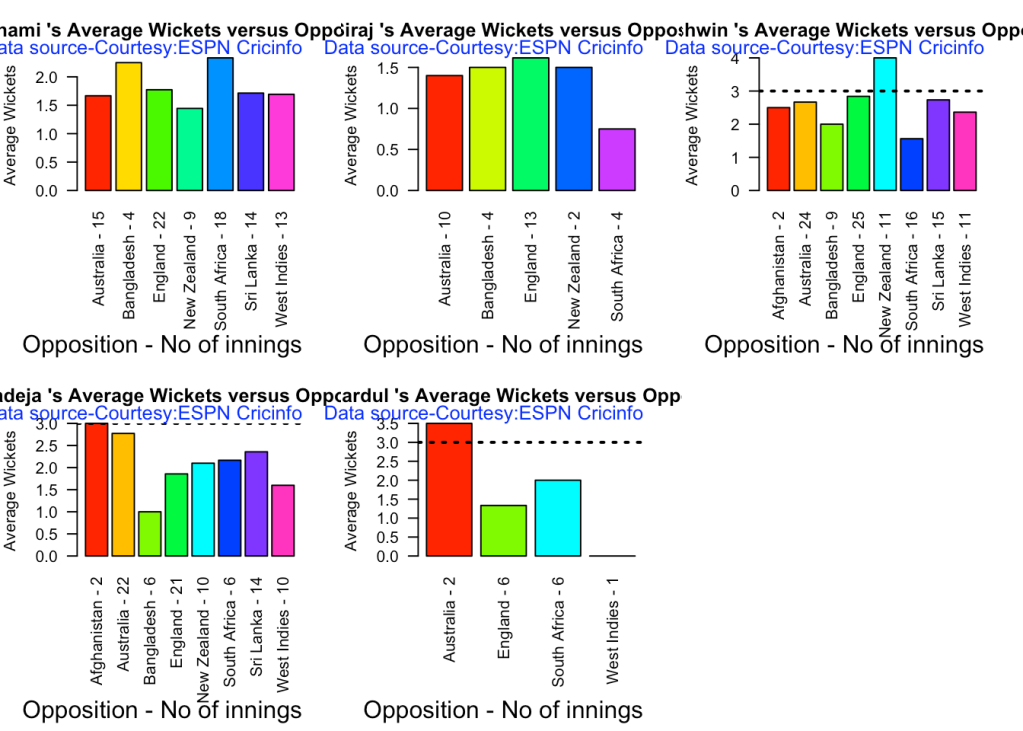
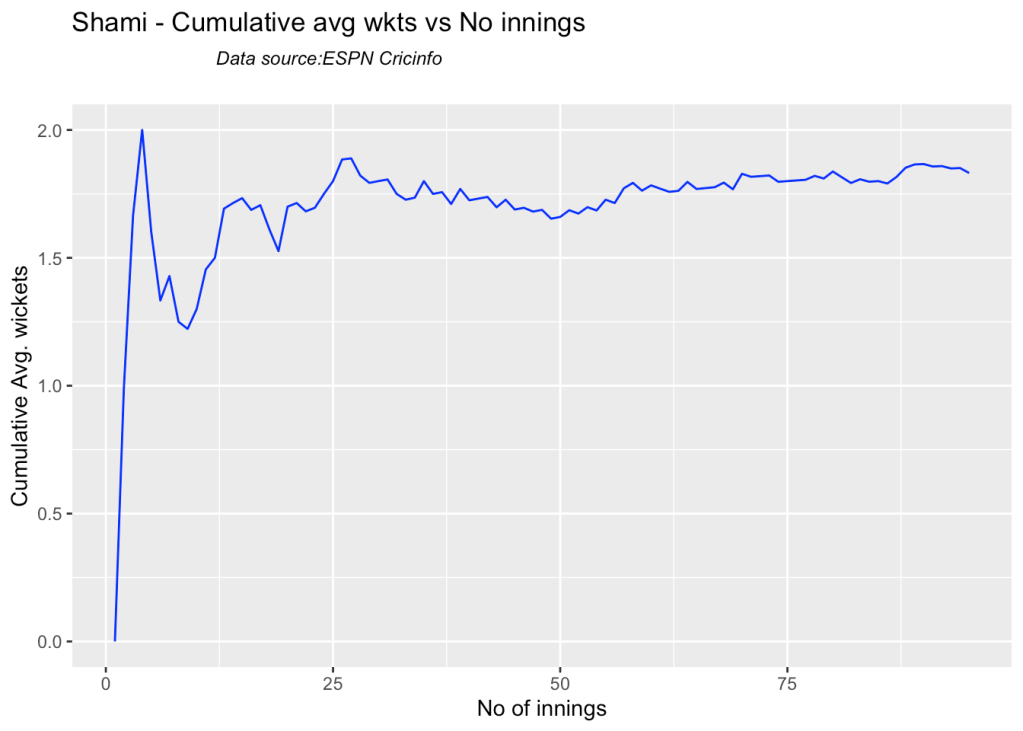
7e Cumulative average wickets taken
Ashwin’s performance has dropped over the years, while Siraj has been becoming better
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
bowlerCumulativeAvgWickets("shamiTest.csv","Shami")
bowlerCumulativeAvgWickets("sirajTest.csv","Siraj")

bowlerCumulativeAvgWickets("ashwinTest.csv","Ashwin")

bowlerCumulativeAvgWickets("jadejaTest.csv","Jadeja")

bowlerCumulativeAvgWickets("shardulTest.csv","Shardul")

7g Cumulative average economy rate
par(mfrow=c(2,3))
par(mar=c(4,4,2,2))
bowlerCumulativeAvgEconRate("shamiTest.csv","Shami")

bowlerCumulativeAvgEconRate("sirajTest.csv","Siraj")

bowlerCumulativeAvgEconRate("ashwinTest.csv","Ashwin")
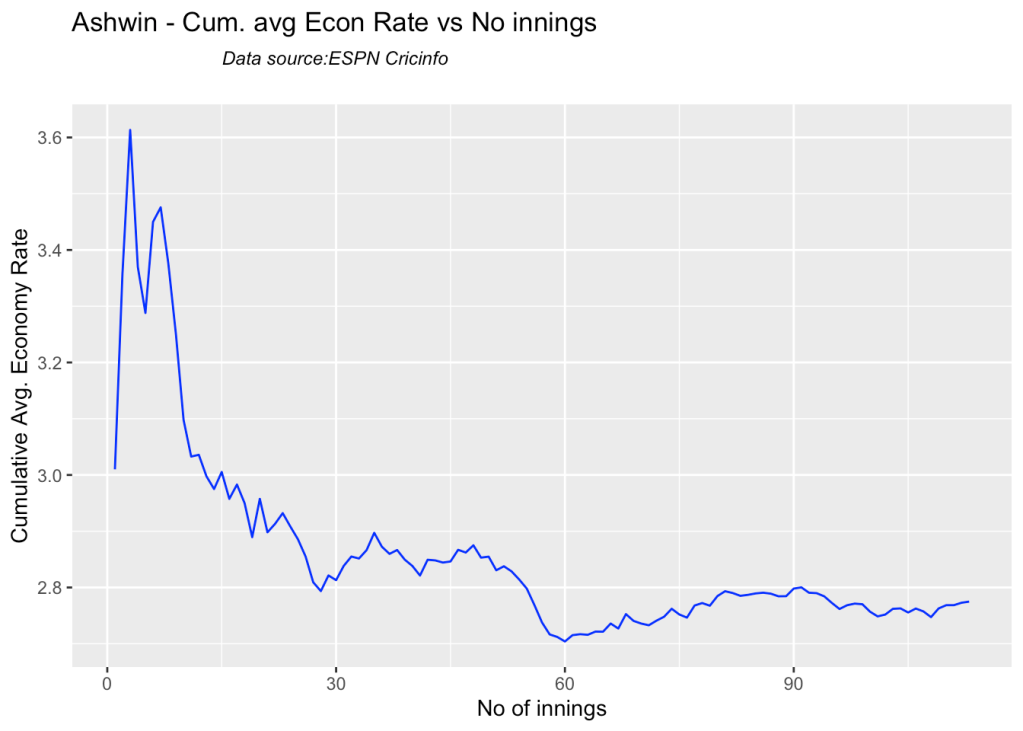
bowlerCumulativeAvgEconRate("jadejaTest.csv","Jadeja")

bowlerCumulativeAvgEconRate("shardulTest.csv","Shardul")

7h Wicket forecast
Here are plots that forecast how the bowler will perform in future. In this case 90% of the career wickets trend is used as the training set. the remaining 10% is the test set.
A Holt-Winters forecasting model is used to forecast future performance based on the 90% training set. The forecasted wickets trend is plotted. The test set is also plotted to see how close the forecast and the actual matches
par(mfrow=c(2,3))
par(mar=c(4,4,2,2))
bowlerPerfForecast("shamiTest.csv","Shami")
#bowlerPerfForecast("sirajTest.csv","Siraj")
bowlerPerfForecast("ashwinTest.csv","Ashwin")
bowlerPerfForecast("jadejaTest.csv","Jadeja")
bowlerPerfForecast("shardulTest.csv","Shardul")

7i Relative Wickets Frequency Percentage
frames <- list("shamiTest.csv","sirajTest.csv","ashwinTest.csv","jadejaTest.csv","shardulTest.csv")
names <- list("Shami","Siraj","Ashwin","Jadeja","Shardul")
relativeBowlingPerf(frames,names)

7j Relative Economy Rate against wickets taken
frames <- list("shamiTest.csv","sirajTest.csv","ashwinTest.csv","jadejaTest.csv","shardulTest.csv")
names <- list("Shami","Siraj","Ashwin","Jadeja","Shardul")
relativeBowlingER(frames,names)

7k Relative cumulative average wickets of bowlers in career
Ashwin has the highest wickets followed by Jadeja against all teams
frames <- list("shamiTest.csv","sirajTest.csv","ashwinTest.csv","jadejaTest.csv","shardulTest.csv")
names <- list("Shami","Siraj","Ashwin","Jadeja","Shardul")
relativeBowlerCumulativeAvgWickets(frames,names)

7l Relative cumulative average economy rate of bowlers
Jadeja has the best economy rate followed by Ashwin
frames <- list("shamiTest.csv","sirajTest.csv","ashwinTest.csv","jadejaTest.csv","shardulTest.csv")
names <- list("Shami","Siraj","Ashwin","Jadeja","Shardul")
relativeBowlerCumulativeAvgEconRate(frames,names)

7m Check for bowler in-form/out-of-form
The below computation uses Null Hypothesis testing and p-value to determine if the bowler is in-form or out-of-form. For this 90% of the career wickets is chosen as the population and the mean computed. The last 10% is chosen to be the sample set and the sample Mean and the sample Standard Deviation are caculated.
The Null Hypothesis (H0) assumes that the bowler continues to stay in-form where the sample mean is within 95% confidence interval of population mean The Alternative (Ha) assumes that the bowler is out of form the sample mean is beyond the 95% confidence interval of the population mean.
A significance value of 0.05 is chosen and p-value us computed If p-value >= .05 – Batsman In-Form If p-value < 0.05 – Batsman Out-of-Form
Note Ideally the p-value should be done for a population that follows the Normal Distribution. But the runs population is usually left skewed. So some correction may be needed. I will revisit this later
Note: The check for the form status of the bowlers indicate
checkBowlerInForm("shami.csv","Shami")
## [1] "**************************** Form status of Shami ****************************\n\n Population size: 106 Mean of population: 1.93 \n Sample size: 12 Mean of sample: 1.33 SD of sample: 1.23 \n\n Null hypothesis H0 : Shami 's sample average is within 95% confidence interval \n of population average\n Alternative hypothesis Ha : Shami 's sample average is below the 95% confidence\n interval of population average\n\n Shami 's Form Status: In-Form because the p value: 0.058427 is greater than alpha= 0.05 \n *******************************************************************************************\n\n"
checkBowlerInForm("siraj.csv","Siraj")
## [1] "**************************** Form status of Siraj ****************************\n\n Population size: 29 Mean of population: 1.59 \n Sample size: 4 Mean of sample: 0.25 SD of sample: 0.5 \n\n Null hypothesis H0 : Siraj 's sample average is within 95% confidence interval \n of population average\n Alternative hypothesis Ha : Siraj 's sample average is below the 95% confidence\n interval of population average\n\n Siraj 's Form Status: Out-of-Form because the p value: 0.002923 is less than alpha= 0.05 \n *******************************************************************************************\n\n"
checkBowlerInForm("ashwin.csv","Ashwin")
## [1] "**************************** Form status of Ashwin ****************************\n\n Population size: 154 Mean of population: 2.77 \n Sample size: 18 Mean of sample: 2.44 SD of sample: 1.76 \n\n Null hypothesis H0 : Ashwin 's sample average is within 95% confidence interval \n of population average\n Alternative hypothesis Ha : Ashwin 's sample average is below the 95% confidence\n interval of population average\n\n Ashwin 's Form Status: In-Form because the p value: 0.218345 is greater than alpha= 0.05 \n *******************************************************************************************\n\n"
checkBowlerInForm("jadeja.csv","Jadeja")
## [1] "**************************** Form status of Jadeja ****************************\n\n Population size: 108 Mean of population: 2.22 \n Sample size: 12 Mean of sample: 1.92 SD of sample: 2.35 \n\n Null hypothesis H0 : Jadeja 's sample average is within 95% confidence interval \n of population average\n Alternative hypothesis Ha : Jadeja 's sample average is below the 95% confidence\n interval of population average\n\n Jadeja 's Form Status: In-Form because the p value: 0.333095 is greater than alpha= 0.05 \n *******************************************************************************************\n\n"
checkBowlerInForm("shardul.csv","Shardul")
## [1] "**************************** Form status of Shardul ****************************\n\n Population size: 13 Mean of population: 2 \n Sample size: 2 Mean of sample: 0.5 SD of sample: 0.71 \n\n Null hypothesis H0 : Shardul 's sample average is within 95% confidence interval \n of population average\n Alternative hypothesis Ha : Shardul 's sample average is below the 95% confidence\n interval of population average\n\n Shardul 's Form Status: Out-of-Form because the p value: 0.04807 is less than alpha= 0.05 \n *******************************************************************************************\n\n"
8. Analysis of India WTC bowlers from Jan 2016 – May 2023 against Australia
8a Relative cumulative average wickets of bowlers in career
Against Australia specifically Jadeja has the best record followed by Ashwin
frames <- list("shamiTestAus.csv","sirajTestAus.csv","ashwinTestAus.csv","jadejaTestAus.csv","shardulTestAus.csv")
names <- list("Shami","Siraj","Ashwin","Jadeja","Shardul")
relativeBowlerCumulativeAvgWickets(frames,names)

8b Relative cumulative average economy rate of bowlers
Jadeja has the best economy followed by Siraj, then Ashwin
frames <- list("shamiTestAus.csv","sirajTestAus.csv","ashwinTestAus.csv","jadejaTestAus.csv","shardulTestAus.csv")
names <- list("Shami","Siraj","Ashwin","Jadeja","Shardul")
relativeBowlerCumulativeAvgEconRate(frames,names)

8. Analysis of India WTC bowlers from Jan 2016 – May 2023
8a. Wickets frequency chart
par(mfrow=c(2,3))
par(mar=c(4,4,2,2))
bowlerWktsFreqPercent("cumminsTest.csv","Cummins")
bowlerWktsFreqPercent("starcTest.csv","Starc")
bowlerWktsFreqPercent("hazzlewoodTest.csv","Hazzlewood")
bowlerWktsFreqPercent("todd.csv","Todd")
bowlerWktsFreqPercent("lyonTest.csv","N Lyon")

8b. Wickets frequency chart
par(mfrow=c(2,3))
par(mar=c(4,4,2,2))
bowlerWktsRunsPlot("cumminsTest.csv","Cummins")
bowlerWktsRunsPlot("starcTest.csv","Starc")
bowlerWktsRunsPlot("hazzlewoodTest.csv","Hazzlewood")
bowlerWktsRunsPlot("todd.csv","Todd")
bowlerWktsRunsPlot("lyonTest.csv","N Lyon")
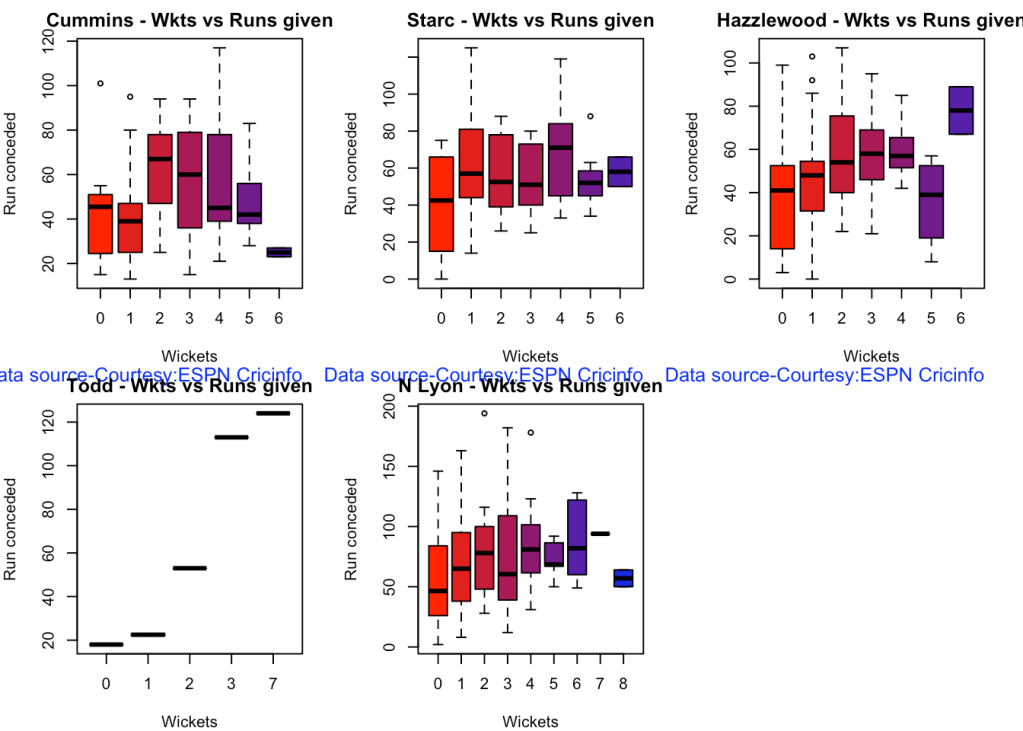
8c. Average wickets at different venues
par(mfrow=c(2,3))
par(mar=c(4,4,2,2))
bowlerAvgWktsGround("cumminsTest.csv","Cummins")
bowlerAvgWktsGround("starcTest.csv","Starc")
bowlerAvgWktsGround("hazzlewoodTest.csv","Hazzlewood")
bowlerAvgWktsGround("todd.csv","Todd")
bowlerAvgWktsGround("lyonTest.csv","N Lyon")

8d Average wickets against different opposition
par(mfrow=c(2,3))
par(mar=c(4,4,2,2))
bowlerAvgWktsOpposition("cumminsTest.csv","Cummins")
bowlerAvgWktsOpposition("starcTest.csv","Starc")
bowlerAvgWktsOpposition("hazzlewoodTest.csv","Hazzlewood")
bowlerAvgWktsOpposition("todd.csv","Todd")
bowlerAvgWktsOpposition("lyonTest.csv","N Lyon")

8e Cumulative average wickets taken
par(mfrow=c(2,2))
par(mar=c(4,4,2,2))
bowlerCumulativeAvgWickets("cumminsTest.csv","Cummins")

bowlerCumulativeAvgWickets("starcTest.csv","Starc")

bowlerCumulativeAvgWickets("hazzlewoodTest.csv","Hazzlewood")

bowlerCumulativeAvgWickets("todd.csv","Todd")

bowlerCumulativeAvgWickets("lyonTest.csv","N Lyon")

8g Cumulative average economy rate
par(mfrow=c(2,3))
par(mar=c(4,4,2,2))
bowlerCumulativeAvgEconRate("cumminsTest.csv","Cummins")

bowlerCumulativeAvgEconRate("starcTest.csv","Starc")

bowlerCumulativeAvgEconRate("hazzlewoodTest.csv","Hazzlewood")

bowlerCumulativeAvgEconRate("todd.csv","Todd")

bowlerCumulativeAvgEconRate("lyonTest.csv","N Lyon")
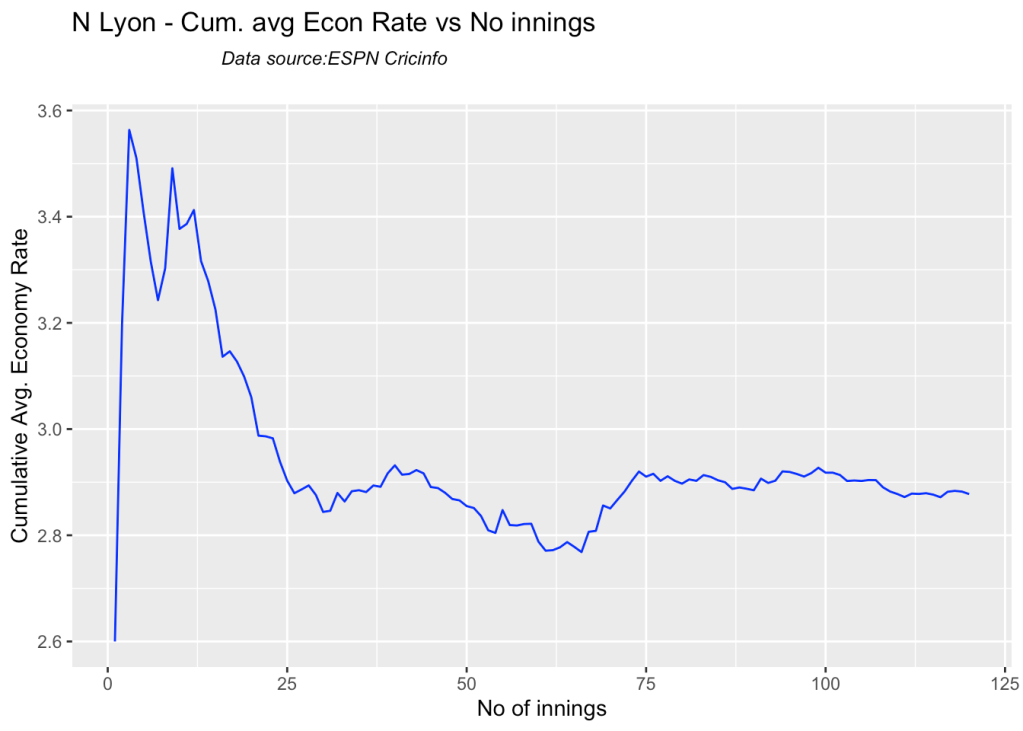
8f. Future Wickets forecast
Here are plots that forecast how the bowler will perform in future. In this case 90% of the career wickets trend is used as the training set. the remaining 10% is the test set.
A Holt-Winters forecasting model is used to forecast future performance based on the 90% training set. The forecated wickets trend is plotted. The test set is also plotted to see how close the forecast and the actual matches
par(mfrow=c(2,3))
par(mar=c(4,4,2,2))
bowlerPerfForecast("cumminsTest.csv","Cummins")
bowlerPerfForecast("starcTest.csv","Starc")
bowlerPerfForecast("hazzlewoodTest.csv","Hazzlewood")
bowlerPerfForecast("lyonTest.csv","N Lyon")

8i. Relative Wickets Frequency Percentage
frames <- list("cumminsTest.csv","starcTest.csv","hazzlewoodTest.csv","todd.csv","lyonTest.csv")
names <- list("Cummins","Starc","Hazzlewood","Todd","N Lyon")
relativeBowlingPerf(frames,names)
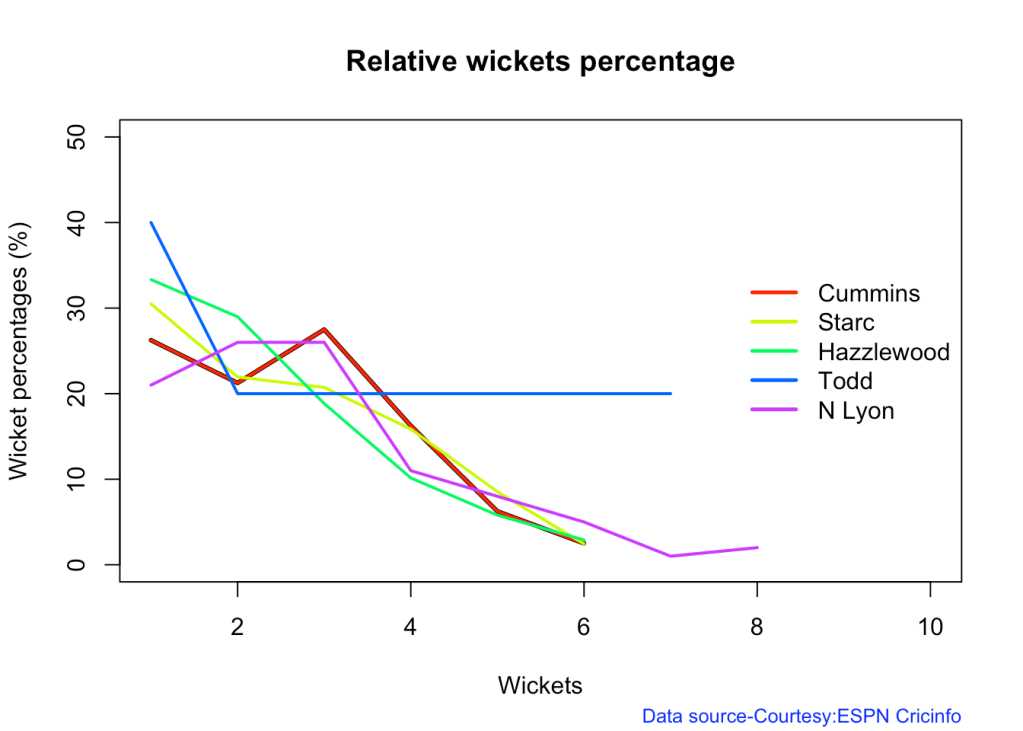
8j Relative Economy Rate against wickets taken
frames <- list("cumminsTest.csv","starcTest.csv","hazzlewoodTest.csv","todd.csv","lyonTest.csv")
names <- list("Cummins","Starc","Hazzlewood","Todd","N Lyon")
relativeBowlingER(frames,names)

8k Relative cumulative average wickets of bowlers in career
Cummins, Starc and Lyons are the best performers
frames <- list("cumminsTest.csv","starcTest.csv","hazzlewoodTest.csv","todd.csv","lyonTest.csv")
names <- list("Cummins","Starc","Hazzlewood","Todd","N Lyon")
relativeBowlerCumulativeAvgWickets(frames,names)
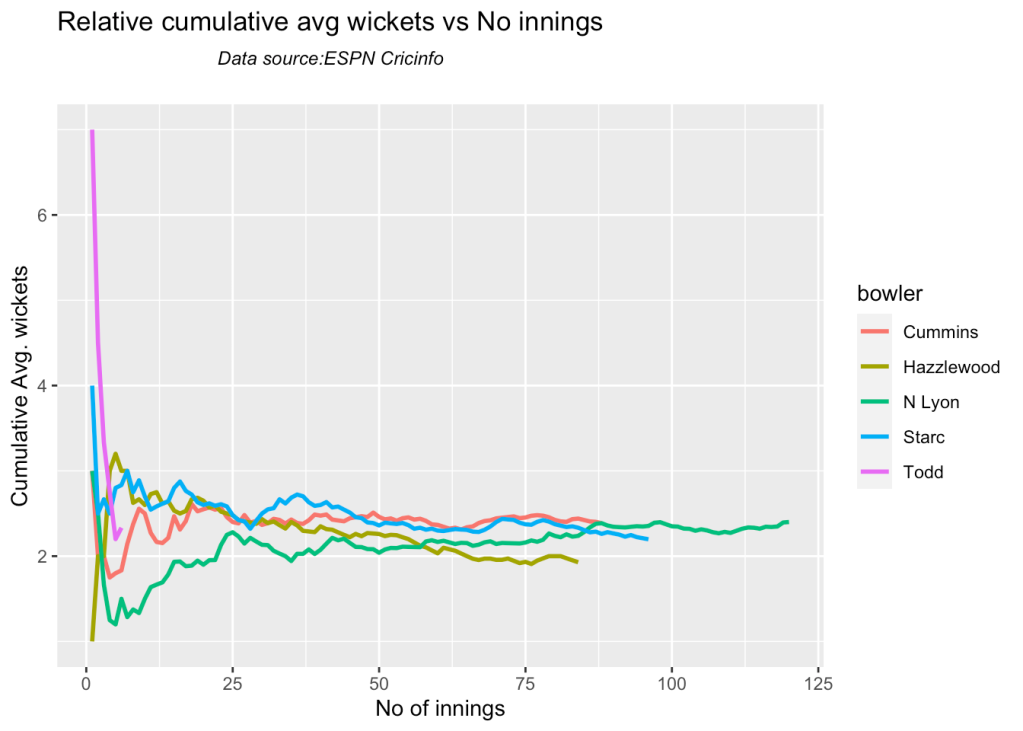
8l Relative cumulative average economy rate of bowlers
Hazzlewood, Cummins have the best economy against all oppostion
frames <- list("cumminsTest.csv","starcTest.csv","hazzlewoodTest.csv","todd.csv","lyonTest.csv")
names <- list("Cummins","Starc","Hazzlewood","Todd","N Lyon")
relativeBowlerCumulativeAvgEconRate(frames,names)

8o Check for bowler in-form/out-of-form
The below computation uses Null Hypothesis testing and p-value to determine if the bowler is in-form or out-of-form. For this 90% of the career wickets is chosen as the population and the mean computed. The last 10% is chosen to be the sample set and the sample Mean and the sample Standard Deviation are calculated.
The Null Hypothesis (H0) assumes that the bowler continues to stay in-form where the sample mean is within 95% confidence interval of population mean The Alternative (Ha) assumes that the bowler is out of form the sample mean is beyond the 95% confidence interval of the population mean.
A significance value of 0.05 is chosen and p-value us computed If p-value >= .05 – Batsman In-Form If p-value < 0.05 – Batsman Out-of-Form
Note Ideally the p-value should be done for a population that follows the Normal Distribution. But the runs population is usually left skewed. So some correction may be needed. I will revisit this later
Note: The check for the form status of the bowlers indicate
checkBowlerInForm("cummins.csv","Cummins")
## [1] "**************************** Form status of Cummins ****************************\n\n Population size: 81 Mean of population: 2.46 \n Sample size: 9 Mean of sample: 2 SD of sample: 1.5 \n\n Null hypothesis H0 : Cummins 's sample average is within 95% confidence interval \n of population average\n Alternative hypothesis Ha : Cummins 's sample average is below the 95% confidence\n interval of population average\n\n Cummins 's Form Status: In-Form because the p value: 0.190785 is greater than alpha= 0.05 \n *******************************************************************************************\n\n"
checkBowlerInForm("starc.csv","Starc")
## [1] "**************************** Form status of Starc ****************************\n\n Population size: 126 Mean of population: 2.18 \n Sample size: 15 Mean of sample: 1.67 SD of sample: 1.18 \n\n Null hypothesis H0 : Starc 's sample average is within 95% confidence interval \n of population average\n Alternative hypothesis Ha : Starc 's sample average is below the 95% confidence\n interval of population average\n\n Starc 's Form Status: In-Form because the p value: 0.057433 is greater than alpha= 0.05 \n *******************************************************************************************\n\n"
checkBowlerInForm("hazzlewood.csv","Hazzlewood")
## [1] "**************************** Form status of Hazzlewood ****************************\n\n Population size: 99 Mean of population: 2.04 \n Sample size: 12 Mean of sample: 1.67 SD of sample: 1.5 \n\n Null hypothesis H0 : Hazzlewood 's sample average is within 95% confidence interval \n of population average\n Alternative hypothesis Ha : Hazzlewood 's sample average is below the 95% confidence\n interval of population average\n\n Hazzlewood 's Form Status: In-Form because the p value: 0.204787 is greater than alpha= 0.05 \n *******************************************************************************************\n\n"
checkBowlerInForm("lyon.csv","N Lyon")
## [1] "**************************** Form status of N Lyon ****************************\n\n Population size: 193 Mean of population: 2.08 \n Sample size: 22 Mean of sample: 2.95 SD of sample: 1.96 \n\n Null hypothesis H0 : N Lyon 's sample average is within 95% confidence interval \n of population average\n Alternative hypothesis Ha : N Lyon 's sample average is below the 95% confidence\n interval of population average\n\n N Lyon 's Form Status: In-Form because the p value: 0.975407 is greater than alpha= 0.05 \n *******************************************************************************************\n\n"
9. Analysis of Australia WTC bowlers from Jan 2016 – May 2023 against India
9a Relative cumulative average wickets of bowlers in career
Against India Lyon, Cummins and Hazzlewood have performed well
frames <- list("cumminsTestInd.csv","starcTestInd.csv","hazzlewoodTestInd.csv","lyonTestInd.csv")
names <- list("Cummins","Starc","Hazzlewood","N Lyon")
relativeBowlerCumulativeAvgWickets(frames,names)

9b Relative cumulative average economy rate of bowlers
Hazzlewood, Lyon have a good economy rate against India
frames <- list("cumminsTestInd.csv","starcTestInd.csv","hazzlewoodTestInd.csv","lyonTestInd.csv")
names <- list("Cummins","Starc","Hazzlewood","N Lyon")
relativeBowlerCumulativeAvgEconRate(frames,names)

10 Analysis of teams – India, Australia
#The data for India & Australia teams were obtained with the following calls
#indiaTest <-getTeamDataHomeAway(dir=".",teamView="bat",matchType="Test",file="indiaTest.csv",save=TRUE,teamName="India")
#australiaTest <- getTeamDataHomeAway(matchType="Test",file="australiaTest.csv",save=TRUE,teamName="Australia")
10a. Win-loss of India against all oppositions in Test cricket
Against Australia India has won 17 times, lost 60 and drawn 22 in Australia. At home India won 42, tied 2, lost 28 and drawn 24
teamWinLossStatusVsOpposition("indiaTest.csv",teamName="India",opposition=c("all"),homeOrAway=c("all"),matchType="Test",plot=TRUE)

10b. Win-loss of Australia against all oppositions in Test cricket
teamWinLossStatusVsOpposition("australiaTest.csv",teamName="Australia",opposition=c("all"),homeOrAway=c("all"),matchType="Test",plot=TRUE)

10c. Win-loss of India against Australia in Test cricket
Against Australia India has won 17 times, lost 60 and drawn 22 in Australia. At home India won 42, tied 2, lost 28 and drawn 24
teamWinLossStatusVsOpposition("indiaTest.csv",teamName="India",opposition=c("Australia"),homeOrAway=c("all"),matchType="Test",plot=TRUE)

10d. Win-loss of India at all away venues
At the Oval where WTC is going to be held India has won 4, lost 10 and drawn 10.
teamWinLossStatusAtGrounds("indiaTest.csv",teamName="India",opposition=c("all"),homeOrAway=c("away"),matchType="Test",plot=TRUE)

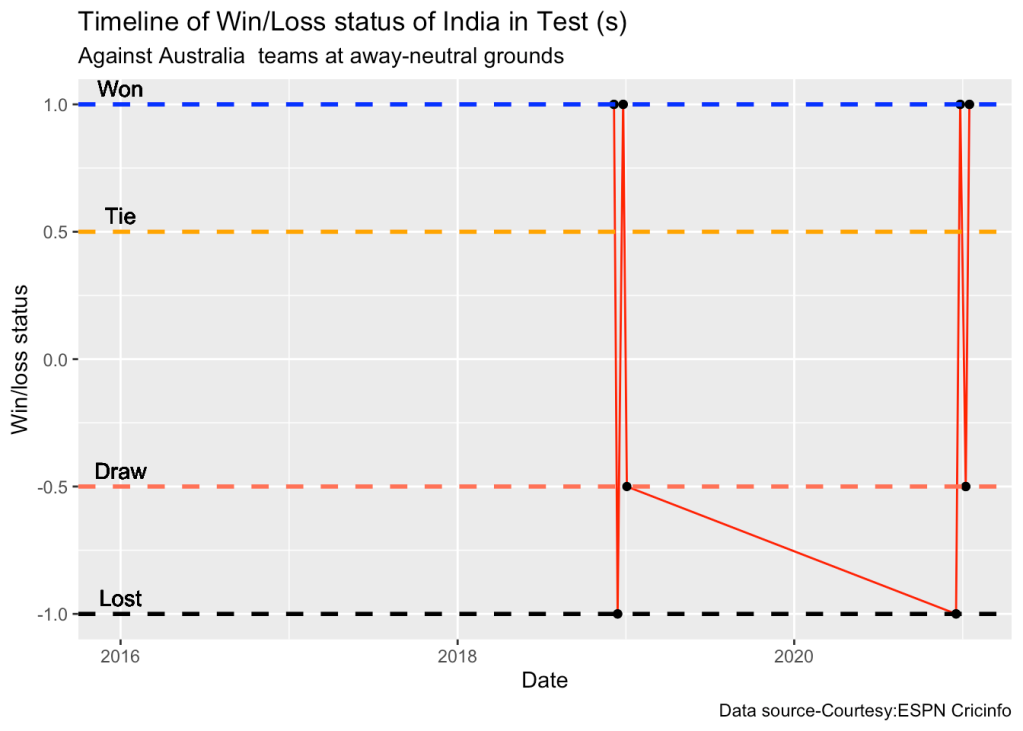
10d. Timeline of win-loss of India against Australia in Test cricket
plotTimelineofWinsLosses("indiaTest.csv",team="India",opposition=c("Australia"),
homeOrAway=c("away","neutral"), startDate="2016-01-01",endDate="2023-05-01")
11. Conclusion
The above analysis performs various analysis of India and Australia in home and away matches. While we know the performance of the player at India or Australia, we cannot judge how the match will progress in the neutral, swinging conditions of the Oval. Let us hope for a good match!
Feel free to try out your own analysis with cricketr. Have fun with cricketr!!
Also see
- GooglyPlusPlus: Win Probability using Deep Learning and player embeddings
- The common alphabet of programming languages
- Practical Machine Learning with R and Python – Part 5
- Deep Learning from first principles in Python, R and Octave – Part 4
- Big Data-4: Webserver log analysis with RDDs, Pyspark, SparkR and SparklyR
- Cricpy takes guard for the Twenty20s
- Using Reinforcement Learning to solve Gridworld
- Exploring Quantum Gate operations with QCSimulator
To see all posts click Index of posts