In this post I use Weights and Biases’ wandb.ai ‘sweep’ feature, to automatically select the best Deep Learning model out of a set of models created through Grid Search. I chanced upon the Weights and Biases site when I was training and fine-tuning the T5 transformer model, on Kaggle, for my post GenerativeAI:Using T5 Transformer model to summarise Indian Philosophy. During this process Kaggle had requested for a token from wandb.ai.
Out of curiosity, I started to explore this Weights and Biases (W&B) machine learning site and was impressed with the visualisation capabilities of this site. So I decided to give weights and biases a try. It is quite interesting to see the live visualisation features of the site and it is becomes very easy to select the optimal model when we are trying to do a Grid search or Random search through a combination of hyper-parameters.

For this purpose, I used my processed T20 match dataset which I had used to compute the Win Probability of T20 teams. For more details please see my post GooglyPlusPlus: Win Probability using Deep Learning and player embeddings
Searching through high dimensional hyperparameter spaces to find the most performant model can quickly get unwieldy. Hyperparameter sweeps provide an organised and efficient way to automatically search through combinations of hyperparameter values (e.g. learning rate, batch size, epochs, dropout, optimizer type) to find the most optimal values.
Here are the steps
a) Install, import
!pip install wandb -qUimport wandb
from wandb.keras import WandbCallback
wandb.login()import pandas as pd
import numpy as np
from zipfile import ZipFile
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras import regularizers
from pathlib import Path
import matplotlib.pyplot as pltb) Load the dataset
import pandas as pd
import numpy as np
from zipfile import ZipFile
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras import regularizers
df1=pd.read_csv('t20.csv')
print("Shape of dataframe=",df1.shape)
train_dataset = df1.sample(frac=0.8,random_state=0)
test_dataset = df1.drop(train_dataset.index)
train_dataset1 = train_dataset[['batsmanIdx','bowlerIdx','ballNum','ballsRemaining','runs','runRate','numWickets','runsMomentum','perfIndex']]
test_dataset1 = test_dataset[['batsmanIdx','bowlerIdx','ballNum','ballsRemaining','runs','runRate','numWickets','runsMomentum','perfIndex']]
train_dataset1
train_labels = train_dataset.pop('isWinner')
test_labels = test_dataset.pop('isWinner')
train_dataset1
a=train_dataset1.describe()
stats=a.transpose
a
Shape of dataframe= (1359888, 10)
batsmanIdx bowlerIdx ballNum ballsRemaining runs runRate numWickets runsMomentum perfIndex
count 1.087910e+06 1.087910e+06 1.087910e+06 1.087910e+06 1.087910e+06 1.087910e+06 1.087910e+06 1.087910e+06 1.087910e+06
mean 2.561058e+03 1.939449e+03 1.185352e+02 6.001942e+01 8.110290e+01 1.611611e+00 2.604912e+00 2.886850e-01 9.619675e+00
std 1.479446e+03 1.095097e+03 6.934078e+01 3.514725e+01 4.977998e+01 2.983874e+00 2.195410e+00 6.066070e-01 4.602859e+00
min 1.000000e+00 1.000000e+00 1.000000e+00 1.000000e+00 -5.000000e+00 -5.000000e+00 0.000000e+00 3.571429e-02 0.000000e+00
25% 1.230000e+03 9.400000e+02 5.900000e+01 3.000000e+01 4.100000e+01 1.043478e+00 1.000000e+00 1.058824e-01 6.539326e+00
50% 2.492000e+03 1.919000e+03 1.170000e+02 5.900000e+01 7.800000e+01 1.300000e+00 2.000000e+00 1.408451e-01 9.246753e+00
75% 3.868000e+03 2.884000e+03 1.770000e+02 9.000000e+01 1.170000e+02 1.590312e+00 4.000000e+00 2.352941e-01 1.218349e+01
max 5.226000e+03 3.848000e+03 2.860000e+02 1.610000e+02 2.780000e+02 2.510000e+02 1.000000e+01 1.100000e+01 6.600000e+01c) Define the Deep Learning model
import pandas as pd
import numpy as np
from keras.layers import Input, Embedding, Flatten, Dense
from keras.models import Model
from keras.layers import Input, Embedding, Flatten, Dense, Reshape, Concatenate, Dropout
from keras.models import Model
tf.random.set_seed(432)
# create input layers for each of the predictors
batsmanIdx_input = Input(shape=(1,), name='batsmanIdx')
bowlerIdx_input = Input(shape=(1,), name='bowlerIdx')
ballNum_input = Input(shape=(1,), name='ballNum')
ballsRemaining_input = Input(shape=(1,), name='ballsRemaining')
runs_input = Input(shape=(1,), name='runs')
runRate_input = Input(shape=(1,), name='runRate')
numWickets_input = Input(shape=(1,), name='numWickets')
runsMomentum_input = Input(shape=(1,), name='runsMomentum')
perfIndex_input = Input(shape=(1,), name='perfIndex')
# Set the embedding size
no_of_unique_batman=len(df1["batsmanIdx"].unique())
print(no_of_unique_batman)
no_of_unique_bowler=len(df1["bowlerIdx"].unique())
print(no_of_unique_bowler)
embedding_size_bat = no_of_unique_batman ** (1/4)
embedding_size_bwl = no_of_unique_bowler ** (1/4)
# create embedding layer for the categorical predictor
batsmanIdx_embedding = Embedding(input_dim=no_of_unique_batman+1, output_dim=16,input_length=1)(batsmanIdx_input)
batsmanIdx_flatten = Flatten()(batsmanIdx_embedding)
bowlerIdx_embedding = Embedding(input_dim=no_of_unique_bowler+1, output_dim=16,input_length=1)(bowlerIdx_input)
bowlerIdx_flatten = Flatten()(bowlerIdx_embedding)
# concatenate all the predictors
x = keras.layers.concatenate([batsmanIdx_flatten,bowlerIdx_flatten, ballNum_input, ballsRemaining_input, runs_input, runRate_input, numWickets_input, runsMomentum_input, perfIndex_input])
# add hidden layers
x = Dense(64, activation='relu')(x)
x = Dropout(0.1)(x)
x = Dense(32, activation='relu')(x)
x = Dropout(0.1)(x)
x = Dense(16, activation='relu')(x)
x = Dropout(0.1)(x)
x = Dense(8, activation='relu')(x)
x = Dropout(0.1)(x)
# add output layer
output = Dense(1, activation='sigmoid', name='output')(x)
print(output.shape)
# create model
# Initialize a new W&B run
#run = wandb.init(project='t20', group='cricket')
model = Model(inputs=[batsmanIdx_input,bowlerIdx_input, ballNum_input, ballsRemaining_input, runs_input, runRate_input, numWickets_input, runsMomentum_input, perfIndex_input], outputs=output)
model.summary()
# Initialize a new W&B run
run = wandb.init(project='t20', group='cricket')
wandb.init(
# set the wandb project where this run will be logged
project="t20",
# track hyperparameters and run metadata
config={
"learning_rate": 0.02,
"dropout": 0.01,
"batch_size": 1024,
"epochs": 5,
}
)
5226
3848
(None, 1)
Model: "model"
__________________________________________________________________________________________________
Layer (type) Output Shape Param # Connected to
==================================================================================================
batsmanIdx (InputLayer) [(None, 1)] 0 []
bowlerIdx (InputLayer) [(None, 1)] 0 []
embedding (Embedding) (None, 1, 16) 83632 ['batsmanIdx[0][0]']
embedding_1 (Embedding) (None, 1, 16) 61584 ['bowlerIdx[0][0]']
flatten (Flatten) (None, 16) 0 ['embedding[0][0]']
flatten_1 (Flatten) (None, 16) 0 ['embedding_1[0][0]']
ballNum (InputLayer) [(None, 1)] 0 []
ballsRemaining (InputLayer [(None, 1)] 0 []
)
runs (InputLayer) [(None, 1)] 0 []
runRate (InputLayer) [(None, 1)] 0 []
numWickets (InputLayer) [(None, 1)] 0 []
runsMomentum (InputLayer) [(None, 1)] 0 []
perfIndex (InputLayer) [(None, 1)] 0 []
concatenate (Concatenate) (None, 39) 0 ['flatten[0][0]',
'flatten_1[0][0]',
'ballNum[0][0]',
'ballsRemaining[0][0]',
'runs[0][0]',
'runRate[0][0]',
'numWickets[0][0]',
'runsMomentum[0][0]',
'perfIndex[0][0]']
dense (Dense) (None, 64) 2560 ['concatenate[0][0]']
dropout (Dropout) (None, 64) 0 ['dense[0][0]']
dense_1 (Dense) (None, 32) 2080 ['dropout[0][0]']
dropout_1 (Dropout) (None, 32) 0 ['dense_1[0][0]']
dense_2 (Dense) (None, 16) 528 ['dropout_1[0][0]']
dropout_2 (Dropout) (None, 16) 0 ['dense_2[0][0]']
dense_3 (Dense) (None, 8) 136 ['dropout_2[0][0]']
dropout_3 (Dropout) (None, 8) 0 ['dense_3[0][0]']
output (Dense) (None, 1) 9 ['dropout_3[0][0]']
==================================================================================================
Total params: 150529 (588.00 KB)
Trainable params: 150529 (588.00 KB)
Non-trainable params: 0 (0.00 Byte)d) Create a Training script
def get_optimizer(lr=1e-2, optimizer="adam"):
"Select optmizer between adam and sgd with momentum"
if optimizer.lower() == "adam":
return tf.keras.optimizers.Adam(learning_rate=lr)
if optimizer.lower() == "sgd":
return tf.keras.optimizers.SGD(learning_rate=lr, momentum=0.1)
def train(model, batch_size=1024, epochs=10, lr=1e-2, optimizer='adam', log_freq=10):
# Compile model like you usually do.
tf.keras.backend.clear_session()
model.compile(loss="binary_crossentropy",
optimizer=get_optimizer(lr, optimizer),
metrics=["accuracy"])
# callback setup
cbs = [WandbCallback(data_type='auto', log_batch_frequency=None)]
# train the model
history=model.fit([train_dataset1['batsmanIdx'],train_dataset1['bowlerIdx'],train_dataset1['ballNum'],train_dataset1['ballsRemaining'],train_dataset1['runs'],
train_dataset1['runRate'],train_dataset1['numWickets'],train_dataset1['runsMomentum'],train_dataset1['perfIndex']], train_labels, epochs=epochs, batch_size=batch_size,callbacks=cbs,
validation_data = ([test_dataset1['batsmanIdx'],test_dataset1['bowlerIdx'],test_dataset1['ballNum'],test_dataset1['ballsRemaining'],test_dataset1['runs'],
test_dataset1['runRate'],test_dataset1['numWickets'],test_dataset1['runsMomentum'],test_dataset1['perfIndex']],test_labels), verbose=1)e) Define the sweep for Grid Search
#Grid search
sweep_config = {
'method': 'grid'
}
metric = {
'name': 'val_loss',
'goal': 'minimize'
}
sweep_config['metric'] = metric
# Optimizers - Adam, SGD
parameters_dict = {
'optimizer': {
'values': ['adam', 'sgd']
},
'dropout': {
'values': [0.1, 0.05]
},
}
sweep_config['parameters'] = parameters_dict
parameters_dict.update({
'epochs': {
'value': 20}
})
import math
# Set learning_rate, batch_size
parameters_dict.update({
'learning_rate': {
'values': [0.005,0.008,0.01,.03]
},
'batch_size': {
'values': [1024,2048]
}
})
import pprint
pprint.pprint(sweep_config)
'method': 'grid',
'metric': {'goal': 'minimize', 'name': 'val_loss'},
'parameters': {'batch_size': {'values': [1024, 2048]},
'dropout': {'values': [0.1, 0.05]},
'epochs': {'value': 20},
'learning_rate': {'values': [0.005, 0.008, 0.01, 0.03]},
'optimizer': {'values': ['adam', 'sgd']}}}
f) Wrap the Training Loop
def sweep_train(config_defaults=None):
# Initialize wandb with a sample project name
with wandb.init(config=config_defaults): # this gets over-written in the Sweep
# Specify the other hyperparameters to the configuration, if any
wandb.config.architecture_name = "DL"
wandb.config.dataset_name = "T20"
# initialize model
#model = T20Net(wandb.config.dropout)
train(model,
wandb.config.batch_size,
wandb.config.epochs,
wandb.config.learning_rate,
wandb.config.optimizer)g) Initialise Sweep and Run Agent
sweep_id = wandb.sweep(sweep_config, project="sweeps-keras-t20")wandb.agent(sweep_id, sweep_train, count=10)wandb: WARNING Calling wandb.login() after wandb.init() has no effect.
wandb: Agent Starting Run: zbaaq0bn with config:
wandb: batch_size: 1024
wandb: dropout: 0.1
wandb: epochs: 20
wandb: learning_rate: 0.005
wandb: optimizer: adam
Epoch 19/20
1061/1063 [============================>.] - ETA: 0s - loss: 0.3073 - accuracy: 0.8490/usr/local/lib/python3.10/dist-packages/keras/src/engine/training.py:3000: UserWarning: You are saving your model as an HDF5 file via `model.save()`. This file format is considered legacy. We recommend using instead the native Keras format, e.g. `model.save('my_model.keras')`.
saving_api.save_model(
wandb: Adding directory to artifact (/content/wandb/run-20231004_065327-zbaaq0bn/files/model-best)... Done. 0.0s
1063/1063 [==============================] - 15s 14ms/step - loss: 0.3073 - accuracy: 0.8490 - val_loss: 0.3093 - val_accuracy: 0.8479
Epoch 20/20
1062/1063 [============================>.] - ETA: 0s - loss: 0.3052 - accuracy: 0.8502/usr/local/lib/python3.10/dist-packages/keras/src/engine/training.py:3000: UserWarning: You are saving your model as an HDF5 file via `model.save()`. This file format is considered legacy. We recommend using instead the native Keras format, e.g. `model.save('my_model.keras')`.
saving_api.save_model(
wandb: Adding directory to artifact (/content/wandb/run-20231004_065327-zbaaq0bn/files/model-best)... Done. 0.0s
1063/1063 [==============================] - 18s 17ms/step - loss: 0.3052 - accuracy: 0.8502 - val_loss: 0.3068 - val_accuracy: 0.8490
Waiting for W&B process to finish... (success).
Run history:
accuracy ▁▅▅▆▆▆▇▇▇▇▇▇▇███████
epoch ▁▁▂▂▂▃▃▄▄▄▅▅▅▆▆▇▇▇██
loss █▅▄▃▃▃▃▂▂▂▂▂▂▂▁▁▁▁▁▁
val_accuracy ▁▂▃▄▄▅▅▅▆▆▆▇▇▇▇▇▇███
val_loss █▆▅▅▄▄▃▃▃▃▃▂▂▂▂▂▂▁▁▁
Run summary:
accuracy 0.85022
best_epoch 19
best_val_loss 0.30681
epoch 19
loss 0.30521
val_accuracy 0.849
val_loss 0.30681
...
...
wandb: Agent Starting Run: 4qtyxzq9 with config:
wandb: batch_size: 1024
wandb: dropout: 0.1
wandb: epochs: 20
wandb: learning_rate: 0.008
wandb: optimizer: sgd
...
...
Epoch 18/20
1063/1063 [==============================] - 13s 12ms/step - loss: 0.2672 - accuracy: 0.8697 - val_loss: 0.2819 - val_accuracy: 0.8624
Epoch 19/20
1061/1063 [============================>.] - ETA: 0s - loss: 0.2669 - accuracy: 0.8697/usr/local/lib/python3.10/dist-packages/keras/src/engine/training.py:3000: UserWarning: You are saving your model as an HDF5 file via `model.save()`. This file format is considered legacy. We recommend using instead the native Keras format, e.g. `model.save('my_model.keras')`.
saving_api.save_model(
wandb: Adding directory to artifact (/content/wandb/run-20231004_070920-4qtyxzq9/files/model-best)... Done. 0.0s
1063/1063 [==============================] - 14s 13ms/step - loss: 0.2669 - accuracy: 0.8697 - val_loss: 0.2813 - val_accuracy: 0.8635
Epoch 20/20
1063/1063 [==============================] - 13s 12ms/step - loss: 0.2650 - accuracy: 0.8707 - val_loss: 0.2957 - val_accuracy: 0.8557
Waiting for W&B process to finish... (success).
6.805 MB of 6.818 MB uploaded (0.108 MB deduped)
Run history:
accuracy ▁▂▃▃▄▄▄▄▄▄▄▄▅▅▄▆▅▆▆█
epoch ▁▁▂▂▂▃▃▄▄▄▅▅▅▆▆▇▇▇██
loss █▇▆▆▅▅▅▅▅▅▅▄▄▄▄▄▄▃▃▁
val_accuracy ▇▅▅▁█▅▇▆▆▅█▅▅▆▃▇▁▇█▁
val_loss ▃▄▄▅▁▃▂▃▃▃▁▄▄▂▆▂█▁▁█
Run summary:
accuracy 0.87067
best_epoch 18
best_val_loss 0.28127
epoch 19
loss 0.26499
val_accuracy 0.85565
val_loss 0.29573
...
...
wandb: Agent Starting Run: lt2fknva with config:
wandb: batch_size: 1024
wandb: dropout: 0.1
wandb: epochs: 20
wandb: learning_rate: 0.01
wandb: optimizer: adam
Tracking run with wandb version 0.15.11
Run data is saved locally in /content/wandb/run-20231004_071359-lt2fknva
Syncing run lively-sweep-5 to Weights & Biases (docs)
...
...
Epoch 19/20
1063/1063 [==============================] - 14s 13ms/step - loss: 0.2779 - accuracy: 0.8651 - val_loss: 0.2883 - val_accuracy: 0.8607
Epoch 20/20
1060/1063 [============================>.] - ETA: 0s - loss: 0.2795 - accuracy: 0.8643/usr/local/lib/python3.10/dist-packages/keras/src/engine/training.py:3000: UserWarning: You are saving your model as an HDF5 file via `model.save()`. This file format is considered legacy. We recommend using instead the native Keras format, e.g. `model.save('my_model.keras')`.
saving_api.save_model(
wandb: Adding directory to artifact (/content/wandb/run-20231004_071359-lt2fknva/files/model-best)... Done. 0.0s
1063/1063 [==============================] - 16s 15ms/step - loss: 0.2795 - accuracy: 0.8643 - val_loss: 0.2831 - val_accuracy: 0.8620
Waiting for W&B process to finish... (success).
Run history:
accuracy ▁▁▁▂▂▃▃▄▅▅▅▆▆▆▆▆▇▇█▇
epoch ▁▁▂▂▂▃▃▄▄▄▅▅▅▆▆▇▇▇██
loss ███▇▇▆▅▆▅▄▄▃▃▃▃▂▂▂▁▂
val_accuracy ▁▅▂▆▆▅▂▆▆▅▇▇▆▇▅▃▃▆▇█
val_loss ▇▆▇▅▃▅█▆▅▄▂▃▄▂▆▆▇▃▃▁
Run summary:
accuracy 0.8643
best_epoch 19
best_val_loss 0.28309
epoch 19
loss 0.27949
val_accuracy 0.86195
val_loss 0.28309
...
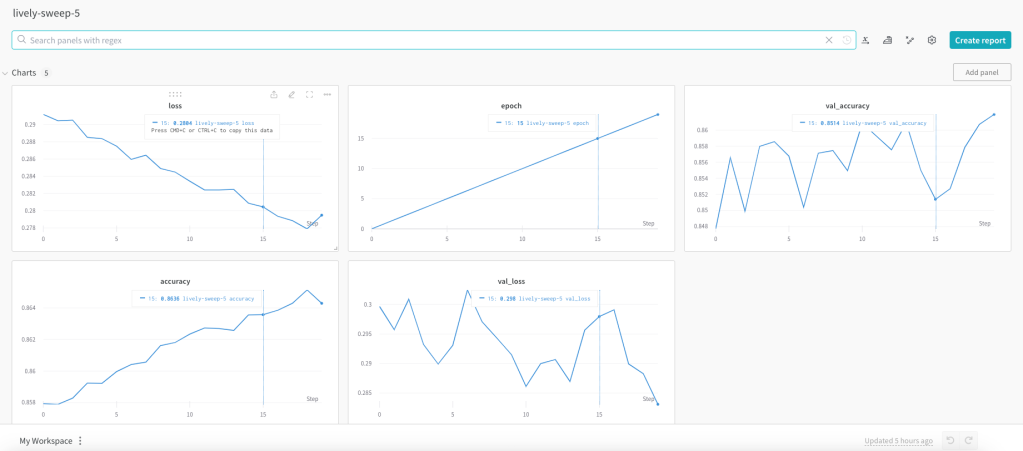
...In the W & B site each of the runs or captured very nicely

The best model is ‘lively-sweep-5‘ with the lowest validation loss

The picture below gives the validation loss for various combinations of the hyper-para meters

It is very easy to visually pick the best model with loss as shown below. It is lively-sweep-5. we can see the values of the hyper-parameters for this DL model
Details of optimal Deep Learning model
a. Run – lively-sweep-5
b. optimizer – adam
c. learning_rate – 0.01
d. batch_size – 1024
e. dropout – 0.1

We can see the performance of this model individually by clicking lively-sweep-6 on the left panel
It was good fun to play around with the Weights and Biases in selecting an optimal model
See also
- Deconstructing Convolutional Neural Networks with Tensorflow and Keras
- Deep Learning from first principles in Python, R and Octave – Part 7
- Identifying cricketing shots using AI
- Introducing cricket package yorkr: Part 2-Trapped leg before wicket!
- Introducing cricpy:A python package to analyze performances of cricketers
To see all posts click Index of posts












































 Checkout my book ‘Deep Learning from first principles: Second Edition – In vectorized Python, R and Octave’. My book starts with the implementation of a simple 2-layer Neural Network and works its way to a generic L-Layer Deep Learning Network, with all the bells and whistles. The derivations have been discussed in detail. The code has been extensively commented and included in its entirety in the Appendix sections. My book is available on Amazon as
Checkout my book ‘Deep Learning from first principles: Second Edition – In vectorized Python, R and Octave’. My book starts with the implementation of a simple 2-layer Neural Network and works its way to a generic L-Layer Deep Learning Network, with all the bells and whistles. The derivations have been discussed in detail. The code has been extensively commented and included in its entirety in the Appendix sections. My book is available on Amazon as 









 Checkout my book ‘Deep Learning from first principles: Second Edition – In vectorized Python, R and Octave’. My book starts with the implementation of a simple 2-layer Neural Network and works its way to a generic L-Layer Deep Learning Network, with all the bells and whistles. The derivations have been discussed in detail. The code has been extensively commented and included in its entirety in the Appendix sections. My book is available on Amazon as
Checkout my book ‘Deep Learning from first principles: Second Edition – In vectorized Python, R and Octave’. My book starts with the implementation of a simple 2-layer Neural Network and works its way to a generic L-Layer Deep Learning Network, with all the bells and whistles. The derivations have been discussed in detail. The code has been extensively commented and included in its entirety in the Appendix sections. My book is available on Amazon as  Checkout my book ‘Deep Learning from first principles: Second Edition – In vectorized Python, R and Octave’. My book starts with the implementation of a simple 2-layer Neural Network and works its way to a generic L-Layer Deep Learning Network, with all the bells and whistles. The derivations have been discussed in detail. The code has been extensively commented and included in its entirety in the Appendix sections. My book is available on Amazon as
Checkout my book ‘Deep Learning from first principles: Second Edition – In vectorized Python, R and Octave’. My book starts with the implementation of a simple 2-layer Neural Network and works its way to a generic L-Layer Deep Learning Network, with all the bells and whistles. The derivations have been discussed in detail. The code has been extensively commented and included in its entirety in the Appendix sections. My book is available on Amazon as