This should be my last post on computing T20 Win Probability. In this post I compute Win Probability using Augmented Data with the help of Conditional Tabular Generative Adversarial Networks (CTGANs).
A.Introduction
I started the computation of T20 match Win Probability in my earlier post
a) ‘Computing Win-Probability of T20 matches‘ where I used
- vanilla Logistic Regression to get an accuracy of 0.67,
- Random Forest with Tidy models gave me an accuracy of 0.737
- Deep Learning with Keras also with 0.73.
This was done without player embeddings
b) Next I used player embeddings for batsmen and bowlers in my post Boosting Win Probability accuracy with player embeddings , and my accuracies improved significantly
- glmnet : accuracy – 0.728 and roc_auc – 0.81
- random forest : accuracy – 0.927 and roc_auc – 0.98
- mlp-dnn :accuracy – 0.762 and roc_auc – 0.854
c) Third I tried using Deep Learning with Keras using player embeddings
- DL network gave an accuracy of 0.8639
This was lightweight and could be easily deployed in my Shiny GooglyPlusPlus app as opposed to the Tidymodel’s Random Forest, which was bulky and slow.
d) Finally I decided to try and improve the accuracy of my Deep Learning Model using Synthetic data. Towards this end, my explorations led me to Conditional Tabular Generative Adversarial Networks (CTGANs). CTGAN are GAN networks that can be used with Tabular data as GAN models are not useful with tabular data. However, the best performance I got for
- DL Keras Model + Synthetic data : accuracy =0.77
The poorer accuracy was because CTGAN requires enormous computing power (GPUs) and RAM. The free version of Colab, Kaggle kept crashing when I tried with even 0.1 % of my 1.2 million dataset size. Finally, I tried with just 0.05% and was able to generate synthetic data. Most likely, it is the small sample size and the smaller number of epochs could be the reason for the poor result. In any case, it was worth trying and this approach would possibly work with sufficient computing resources.
B.Generative Adversarial Networks (GANs)
Generative Adversarial Networks (GANs) was the brain child of Ian Goodfellow who demonstrated it in 2014. GANs are capable of generating synthetic text, tables, images, videos using available data. In Adversarial nets framework, the generative model is pitted against an adversary: a
discriminative model that learns to determine whether a sample is from the model distribution or the
data distribution.
GANs have 2 Deep Neural Networks , the Generator and Discriminator which compete against other
- The Generator (Counterfeiter) takes random noise as input and generates fake images, tables, text. The generator learns to generate plausible data. The generated instances become negative training examples for the discriminator.
- The Discriminator (Police) which tries to distinguish between the real and fake images, text. The discriminator learns to distinguish the generator’s fake data from real data. The discriminator penalises the generator for producing implausible results.
A pictorial representation of the GAN model can be shown below

Theoretically best performance of GANs are supposed to happen when the network reaches the ‘Nash equilibrium‘, i.e. when the Generator produces near fake images and the Discriminator’s loss is f ~0.5 i.e. the discriminator is unable to distinguish between real and fake images.
Note: Though I have mentioned T20 data in the above GAN model, the T20 tabular data is actually used in CTGAN which is slightly different from the above. See Reference 2) below.
C. Conditional Tabular Generative Adversial Networks (CTGANs)
“Modeling the probability distribution of rows in tabular data and generating realistic synthetic data is a non-trivial task. Tabular data usually contains a mix of discrete and continuous columns. Continuous columns may have multiple modes whereas discrete columns are sometimes imbalanced making the modeling difficult.” CTGANs handle these challenges.
I came upon CTGAN after spending some time exploring GANs via blogs, videos etc. For building the model I use real T20 match data. However, CTGAN requires immense raw computing power and a lot of RAM. My initial attempts on Colab, my Mac (12 core, 32GB RAM), took forever before eventually crashing, I switched to Kaggle and used GPUs. Still I was only able to use only a miniscule part of my T20 dataset. My match data has 1.2 million rows, hoanything > 0.05% resulted in Kaggle crashing. Since I was able to use only a fraction, I executed the CTGAN model over several iterations, each iteration with a random 0.05% sample of the dataset. At the end of each iterations I also generate synthetic dataset. Over 12 iterations, I generate close 360K of ‘synthetic‘ T20 match data.
I then augment the 1.2 million rows of ‘real‘ T20 match data with the generated ‘synthetic T20 match data to run my Deep Learning model
D. Executing the CTGAN model
a. Read the real T20 match data
!pip install ctgan
import pandas as pd
import ctgan
from ctgan import CTGAN
from numpy.random import seed
# Read the T20 match data
df = pd.read_csv('/kaggle/input/cricket1/t20.csv')
# Randomly sample 0.05% of the dataset. Note larger datasets cause the algo to crash
train_dataset = df.sample(frac=0.05)
# Print the real T20 match data
print(train_dataset.head(10))
print(train_dataset.shape)
batsmanIdx bowlerIdx ballNum ballsRemaining runs runRate \
363695 3333 432 134 119 153 1.285714
1082839 3881 1180 218 30 93 3.100000
595799 2366 683 187 65 120 1.846154
737614 4490 1381 148 87 144 1.655172
410202 934 1003 19 106 35 1.842105
525627 921 1711 251 1 8 8.000000
657669 4718 1602 130 115 145 1.260870
666461 4309 1989 44 87 38 0.863636
651229 3336 754 30 92 36 1.200000
709892 3048 421 97 28 119 1.226804
numWickets runsMomentum perfIndex isWinner
363695 0 0.092437 18.333333 1
1082839 5 0.200000 4.736842 0
595799 4 0.107692 9.566667 0
737614 1 0.114943 9.130435 1
410202 0 0.103774 20.263158 0
525627 8 3.000000 3.837209 0
657669 0 0.095652 19.555556 0
666461 0 0.126437 9.500000 0
651229 0 0.119565 13.200000 0
709892 3 0.285714 9.814433 1
(59956, 10)b. Run CTGAN model on the real T20 data
import pandas as pd
import ctgan
from ctgan import CTGAN
from numpy.random import seed
from pickle import TRUE
df = pd.read_csv('/kaggle/input/cricket1/t20.csv')
#Specify the categorical features. batsmanIdx & bowlerIdx are player embeddings
categorical_features = ['batsmanIdx','bowlerIdx']
# Create a empty dataframe for synthetic data
df1 = pd.DataFrame()
# Loop for 12 iterations. Minimize generator & discriminator loss
for i in range(12):
print(i)
train_dataset = df.sample(frac=0.05)
seed(33)
ctgan = CTGAN(epochs=20,verbose=True,generator_lr=.001,discriminator_lr=.001,batch_size=1000)
ctgan.fit(train_dataset, categorical_features)
# Generate synthetic data
samples = ctgan.sample(30000)
# Concatenate the synthetic data after each iteration
df1 = pd.concat([df1,samples])
print(samples.head())
print(df1.shape)
# Output the synthetic data to file
df1.to_csv("output1.csv",index=False)
0
Epoch 1, Loss G: 8.3825,Loss D: -0.6159
Epoch 2, Loss G: 3.5117,Loss D: -0.3016
Epoch 3, Loss G: 2.1619,Loss D: -0.5713
Epoch 4, Loss G: 0.9847,Loss D: 0.1010
Epoch 5, Loss G: 0.6198,Loss D: 0.0789
Epoch 6, Loss G: 0.1710,Loss D: 0.0959
Epoch 7, Loss G: 0.3236,Loss D: -0.1554
Epoch 8, Loss G: 0.2317,Loss D: -0.0765
Epoch 9, Loss G: -0.0127,Loss D: 0.0275
Epoch 10, Loss G: 0.1477,Loss D: -0.0353
Epoch 11, Loss G: 0.0997,Loss D: -0.0129
Epoch 12, Loss G: 0.0066,Loss D: -0.0486
Epoch 13, Loss G: 0.0351,Loss D: -0.0805
Epoch 14, Loss G: -0.1399,Loss D: -0.0021
Epoch 15, Loss G: -0.1503,Loss D: -0.0518
Epoch 16, Loss G: -0.2306,Loss D: -0.0234
Epoch 17, Loss G: -0.2986,Loss D: 0.0469
Epoch 18, Loss G: -0.1941,Loss D: -0.0560
Epoch 19, Loss G: -0.3794,Loss D: 0.0000
Epoch 20, Loss G: -0.2763,Loss D: 0.0368
batsmanIdx bowlerIdx ballNum ballsRemaining runs runRate numWickets \
0 906 224 8 75 81 1.955153 4
1 4159 433 17 31 126 1.799280 9
2 229 351 192 66 82 1.608527 5
3 1926 962 63 0 117 1.658105 0
4 286 431 128 1 36 1.605079 0
runsMomentum perfIndex isWinner
0 0.146670 6.937595 1
1 0.160534 10.904346 1
2 0.516010 11.698128 1
3 0.380986 11.914613 0
4 0.112255 5.392120 0
(30000, 10)
1
Epoch 1, Loss G: 7.9977,Loss D: -0.3592
Epoch 2, Loss G: 3.7418,Loss D: -0.3371
Epoch 3, Loss G: 1.6685,Loss D: -0.3211
Epoch 4, Loss G: 1.0539,Loss D: -0.3495
Epoch 5, Loss G: 0.4664,Loss D: -0.0907
Epoch 6, Loss G: 0.4004,Loss D: -0.1208
Epoch 7, Loss G: 0.3250,Loss D: -0.1482
Epoch 8, Loss G: 0.1753,Loss D: 0.0169
Epoch 9, Loss G: 0.1382,Loss D: 0.0661
Epoch 10, Loss G: 0.1509,Loss D: -0.1023
Epoch 11, Loss G: -0.0235,Loss D: 0.0210
Epoch 12, Loss G: -0.1636,Loss D: -0.0124
Epoch 13, Loss G: -0.3370,Loss D: -0.0185
Epoch 14, Loss G: -0.3054,Loss D: -0.0085
Epoch 15, Loss G: -0.5142,Loss D: 0.0121
Epoch 16, Loss G: -0.3813,Loss D: -0.0921
Epoch 17, Loss G: -0.5838,Loss D: 0.0210
Epoch 18, Loss G: -0.4033,Loss D: -0.0181
Epoch 19, Loss G: -0.5711,Loss D: 0.0269
Epoch 20, Loss G: -0.4828,Loss D: -0.0830
batsmanIdx bowlerIdx ballNum ballsRemaining runs runRate numWickets \
0 2202 265 223 39 13 0.868927 0
1 3641 856 35 59 26 2.236160 6
2 676 2903 218 93 16 0.460693 1
3 3482 3459 44 117 102 0.851471 8
4 3046 3076 59 5 84 1.016824 2
runsMomentum perfIndex isWinner
0 0.138586 4.733462 0
1 0.124453 5.146831 1
2 0.273168 10.106869 0
3 0.129520 5.361127 0
4 1.083525 25.677574 1
(60000, 10)
...
...
11
Epoch 1, Loss G: 8.8362,Loss D: -0.7111
Epoch 2, Loss G: 4.1322,Loss D: -0.8468
Epoch 3, Loss G: 1.2782,Loss D: 0.1245
Epoch 4, Loss G: 1.1135,Loss D: -0.3588
Epoch 5, Loss G: 0.6033,Loss D: -0.1255
Epoch 6, Loss G: 0.6912,Loss D: -0.1906
Epoch 7, Loss G: 0.3340,Loss D: -0.1048
Epoch 8, Loss G: 0.3515,Loss D: -0.0730
Epoch 9, Loss G: 0.1702,Loss D: 0.0237
Epoch 10, Loss G: 0.1064,Loss D: 0.0632
Epoch 11, Loss G: 0.0884,Loss D: -0.0005
Epoch 12, Loss G: 0.0556,Loss D: -0.0607
Epoch 13, Loss G: -0.0917,Loss D: -0.0223
Epoch 14, Loss G: -0.1492,Loss D: 0.0258
Epoch 15, Loss G: -0.0986,Loss D: -0.0112
Epoch 16, Loss G: -0.1428,Loss D: -0.0060
Epoch 17, Loss G: -0.2225,Loss D: -0.0263
Epoch 18, Loss G: -0.2255,Loss D: -0.0328
Epoch 19, Loss G: -0.3482,Loss D: 0.0277
Epoch 20, Loss G: -0.2667,Loss D: -0.0721
batsmanIdx bowlerIdx ballNum ballsRemaining runs runRate numWickets \
0 367 1447 129 27 30 1.242120 2
1 2481 1528 221 4 10 1.344024 2
2 1034 3116 132 87 153 1.142750 3
3 1201 2868 151 60 136 1.091638 1
4 4327 3291 108 89 22 0.842775 2
runsMomentum perfIndex isWinner
0 1.978739 6.393691 1
1 0.539650 6.783990 0
2 0.107156 12.154197 0
3 3.193574 11.992059 0
4 0.127507 12.210876 0
(360000, 10)E. Sample of the Synthetic data
synthetic_data = ctgan.sample(20000)
print(synthetic_data.head(100))
batsmanIdx bowlerIdx ballNum ballsRemaining runs runRate \
0 1073 3059 72 72 149 2.230236
1 3769 1443 106 7 137 0.881409
2 448 3048 166 6 220 1.092504
3 2969 1244 103 82 207 12.314862
4 180 1372 125 111 14 1.310051
.. ... ... ... ... ... ...
95 1521 1040 153 6 166 1.097363
96 2366 62 25 114 119 0.910642
97 3506 1736 100 118 140 1.640921
98 3343 2347 47 54 50 0.696462
99 1957 2888 136 27 153 1.315565
numWickets runsMomentum perfIndex isWinner
0 0 0.111707 17.466925 0
1 1 0.130352 14.274113 0
2 1 0.173541 11.076731 1
3 1 0.218977 6.239951 0
4 4 2.829380 9.183323 1
.. ... ... ... ...
95 0 0.223437 7.011180 0
96 1 0.451371 16.908120 1
97 5 0.156936 9.217205 0
98 6 0.124536 6.273091 0
99 1 0.249329 14.221554 0
[100 rows x 10 columns]F. Evaluating the synthetic T20 match data
Here the quality of the synthetic data set is evaluated.
a) Statistical evaluation
- Read the real T20 match data
- Read the generated T20 synthetic match data
import pandas as pd
# Read the T20 match and synthetic match data
df = pd.read_csv('/kaggle/input/cricket1/t20.csv'). #1.2 million rows
synthetic=pd.read_csv('/kaggle/input/synthetic/synthetic.csv') #300K
# Randomly sample 1000 rows, and generate stats
df1=df.sample(n=1000)
real=df1.describe()
realData_stats=real.transpose
print(realData_stats)
synthetic1=synthetic.sample(n=1000)
synthetic=synthetic1.describe()
syntheticData_stats=synthetic.transpose
syntheticData_statsa) Stats of real T20 match data
<bound method DataFrame.transpose of batsmanIdx bowlerIdx ballNum ballsRemaining runs \
count 1000.000000 1000.000000 1000.000000 1000.000000 1000.000000
mean 2323.940000 1776.481000 118.165000 59.236000 77.649000
std 1329.703046 1011.470703 70.564291 35.312934 49.098763
min 8.000000 13.000000 1.000000 1.000000 -2.000000
25% 1134.750000 850.000000 58.000000 28.750000 39.000000
50% 2265.000000 1781.500000 117.000000 59.000000 72.000000
75% 3510.000000 2662.250000 178.000000 89.000000 111.000000
max 4738.000000 3481.000000 265.000000 127.000000 246.000000
runRate numWickets runsMomentum perfIndex isWinner
count 1000.000000 1000.000000 1000.000000 1000.000000 1000.000000
mean 1.734979 2.614000 0.310568 9.580386 0.499000
std 5.698104 2.267189 0.686171 4.530856 0.500249
min -2.000000 0.000000 0.071429 0.000000 0.000000
25% 1.009063 1.000000 0.105769 6.666667 0.000000
50% 1.272727 2.000000 0.141026 9.236842 0.000000
75% 1.546891 4.000000 0.250000 12.146735 1.000000
max 166.000000 10.000000 10.000000 30.800000 1.000000b) Stats of Synthetic T20 match data
batsmanIdx bowlerIdx ballNum ballsRemaining runs \
count 1000.000000 1000.000000 1000.000000 1000.000000 1000.000000
mean 2304.135000 1760.776000 116.081000 50.102000 74.357000
std 1342.348684 1003.496003 72.019228 35.795236 48.103446
min 2.000000 15.000000 -4.000000 -2.000000 -1.000000
25% 1093.000000 881.000000 46.000000 18.000000 30.000000
50% 2219.500000 1763.500000 116.000000 45.000000 75.000000
75% 3496.500000 2644.750000 180.250000 77.000000 112.000000
max 4718.000000 3481.000000 253.000000 124.000000 222.000000
runRate numWickets runsMomentum perfIndex isWinner
count 1000.000000 1000.000000 1000.000000 1000.000000 1000.000000
mean 1.637225 3.096000 0.336540 9.278073 0.507000
std 1.691060 2.640408 0.502346 4.727677 0.500201
min -4.388339 0.000000 0.083351 -0.902991 0.000000
25% 1.077789 1.000000 0.115770 5.731931 0.000000
50% 1.369655 2.000000 0.163085 9.104328 1.000000
75% 1.660477 5.000000 0.311586 12.619318 1.000000
max 23.757001 10.000000 4.630908 29.829497 1.000000c) Plotting the Generator and Discriminator loss
import pandas as pd
# CTGAN prints out a new line for each epoch
epochs_output = str(output).split('\n')
# CTGAN separates the values with commas
raw_values = [line.split(',') for line in epochs_output]
loss_values = pd.DataFrame(raw_values)[:-1] # convert to df and delete last row (empty)
# Rename columns
loss_values.columns = ['Epoch', 'Generator Loss', 'Discriminator Loss']
# Extract the numbers from each column
loss_values['Epoch'] = loss_values['Epoch'].str.extract('(\d+)').astype(int)
loss_values['Generator Loss'] = loss_values['Generator Loss'].str.extract('([-+]?\d*\.\d+|\d+)').astype(float)
loss_values['Discriminator Loss'] = loss_values['Discriminator Loss'].str.extract('([-+]?\d*\.\d+|\d+)').astype(float)
# the result is a row for each epoch that contains the generator and discriminator loss
loss_values.head() Epoch Generator Loss Discriminator Loss
0 1 8.0158 -0.3840
1 2 4.6748 -0.9589
2 3 1.1503 -0.0066
3 4 1.5593 -0.8148
4 5 0.6734 -0.1425
5 6 0.5342 -0.2202
6 7 0.4539 -0.1462
7 8 0.2907 -0.0155
8 9 0.2399 0.0172
9 10 0.1520 -0.0236import plotly.graph_objects as go
# Plot loss function
fig = go.Figure(data=[go.Scatter(x=loss_values['Epoch'], y=loss_values['Generator Loss'], name='Generator Loss'),
go.Scatter(x=loss_values['Epoch'], y=loss_values['Discriminator Loss'], name='Discriminator Loss')])
# Update the layout for best viewing
fig.update_layout(template='plotly_white',
legend_orientation="h",
legend=dict(x=0, y=1.1))
title = 'CTGAN loss function for T20 dataset - '
fig.update_layout(title=title, xaxis_title='Epoch', yaxis_title='Loss')
fig.show()
G. Qualitative evaluation of Synthetic data
a) Quality of continuous columns in synthetic data
KSComplement -This metric computes the similarity of a real column vs. a synthetic column in terms of the column shapes.The KSComplement uses the Kolmogorov-Smirnov statistic. Closer to 1.0 is good and 0 is worst
from sdmetrics.single_column import KSComplement
numerical_columns=['ballNum','ballsRemaining','runs','runRate','numWickets','runsMomentum','perfIndex']
total_score = 0
for column_name in numerical_columns:
column_score = KSComplement.compute(df[column_name], synthetic[column_name])
total_score += column_score
print('Column:', column_name, ', Score: ', column_score)
print('\nAverage: ', total_score/len(numerical_columns))
Column: ballNum , Score: 0.9502754283367316
Column: ballsRemaining , Score: 0.8770284103276166
Column: runs , Score: 0.9136464248633367
Column: runRate , Score: 0.9183841670732166
Column: numWickets , Score: 0.9016209114638712
Column: runsMomentum , Score: 0.8773491702213716
Column: perfIndex , Score: 0.9173808852778924
Average: 0.9079550567948624b) Quality of categorical columns
This statistic measures the quality of generated categorical columns. 1 is best and 0 is worst
categorical_columns=['batsmanIdx','bowlerIdx']
from sdmetrics.single_column import TVComplement
total_score = 0
for column_name in categorical_columns:
column_score = TVComplement.compute(df[column_name], synthetic[column_name])
total_score += column_score
print('Column:', column_name, ', Score: ', column_score)
print('\nAverage: ', total_score/len(categorical_columns))
Column: batsmanIdx , Score: 0.8436263499539245
Column: bowlerIdx , Score: 0.7356177407921669
Average: 0.7896220453730457The performance is decent but not excellent. I was unable to execute more epochs as it it required larger than the memory allowed
c) Correlation similarity
This metric measures the correlation between a pair of numerical columns and computes the similarity between the real and synthetic data – it compares the trends of 2D distributions. Best 1.0 and 0.0 is worst
import itertools
from sdmetrics.column_pairs import CorrelationSimilarity
total_score = 0
total_pairs = 0
for pair in itertools.combinations(numerical_columns,2):
col_A, col_B = pair
score = CorrelationSimilarity.compute(df[[col_A, col_B]], synthetic[[col_A, col_B]])
print('Columns:', pair, ' Score:', score)
total_score += score
total_pairs += 1
print('\nAverage: ', total_score/total_pairs)
Columns: ('ballNum', 'ballsRemaining') Score: 0.7153942317384889
Columns: ('ballNum', 'runs') Score: 0.8838043045134777
Columns: ('ballNum', 'runRate') Score: 0.8710243133637056
Columns: ('ballNum', 'numWickets') Score: 0.7978515509750435
Columns: ('ballNum', 'runsMomentum') Score: 0.8956281260834316
Columns: ('ballNum', 'perfIndex') Score: 0.9275145840528048
Columns: ('ballsRemaining', 'runs') Score: 0.9566928975064546
Columns: ('ballsRemaining', 'runRate') Score: 0.9127313819127167
Columns: ('ballsRemaining', 'numWickets') Score: 0.6770737279315224
Columns: ('ballsRemaining', 'runsMomentum') Score: 0.7939260278412358
Columns: ('ballsRemaining', 'perfIndex') Score: 0.8694582252638351
Columns: ('runs', 'runRate') Score: 0.999593795992159
Columns: ('runs', 'numWickets') Score: 0.9510731832916608
Columns: ('runs', 'runsMomentum') Score: 0.9956131422133428
Columns: ('runs', 'perfIndex') Score: 0.9742931845536701
Columns: ('runRate', 'numWickets') Score: 0.8859830711832263
Columns: ('runRate', 'runsMomentum') Score: 0.9174744874779561
Columns: ('runRate', 'perfIndex') Score: 0.9491100087911353
Columns: ('numWickets', 'runsMomentum') Score: 0.8989709776329797
Columns: ('numWickets', 'perfIndex') Score: 0.7178946968801441
Columns: ('runsMomentum', 'perfIndex') Score: 0.9744441623018661
Average: 0.8840738134048025d) Category coverage
This metric measures whether a synthetic column covers all the possible categories that are present in a real column. 1.0 is best , 0 is worst
from sdmetrics.single_column import CategoryCoverage
total_score = 0
for column_name in categorical_columns:
column_score = CategoryCoverage.compute(df[column_name], synthetic[column_name])
total_score += column_score
print('Column:', column_name, ', Score: ', column_score)
print('\nAverage: ', total_score/len(categorical_columns))
Column: batsmanIdx , Score: 0.9533951919021509
Column: bowlerIdx , Score: 0.9913966160022942
Average: 0.9723959039522225H. Augmenting the T20 match data set
In this final part I augment my T20 match data set with the generated synthetic T20 data set.
import pandas as pd
from numpy import savetxt
import tensorflow as tf
from tensorflow import keras
import pandas as pd
import numpy as np
from keras.layers import Input, Embedding, Flatten, Dense, Reshape, Concatenate, Dropout
from keras.models import Model
import matplotlib.pyplot as plt
# Read real and synthetic data
df = pd.read_csv('/kaggle/input/cricket1/t20.csv')
synthetic=pd.read_csv('/kaggle/input/synthetic/synthetic.csv')
# Augment the data. Concatenate real & synthetic data
df1=pd.concat([df,synthetic])
# Create training and test samples
print("Shape of dataframe=",df1.shape)
train_dataset = df1.sample(frac=0.8,random_state=0)
test_dataset = df1.drop(train_dataset.index)
train_dataset1 = train_dataset[['batsmanIdx','bowlerIdx','ballNum','ballsRemaining','runs','runRate','numWickets','runsMomentum','perfIndex']]
test_dataset1 = test_dataset[['batsmanIdx','bowlerIdx','ballNum','ballsRemaining','runs','runRate','numWickets','runsMomentum','perfIndex']]
train_dataset1
train_labels = train_dataset.pop('isWinner')
test_labels = test_dataset.pop('isWinner')
print(train_dataset1.shape)
a=train_dataset1.describe()
stats=a.transpose
print(a)a) Create A Deep Learning Model in Keras
from numpy.random import seed
seed(33)
tf.random.set_seed(432)
# create input layers for each of the predictors
batsmanIdx_input = Input(shape=(1,), name='batsmanIdx')
bowlerIdx_input = Input(shape=(1,), name='bowlerIdx')
ballNum_input = Input(shape=(1,), name='ballNum')
ballsRemaining_input = Input(shape=(1,), name='ballsRemaining')
runs_input = Input(shape=(1,), name='runs')
runRate_input = Input(shape=(1,), name='runRate')
numWickets_input = Input(shape=(1,), name='numWickets')
runsMomentum_input = Input(shape=(1,), name='runsMomentum')
perfIndex_input = Input(shape=(1,), name='perfIndex')
no_of_unique_batman=len(df1["batsmanIdx"].unique())
print(no_of_unique_batman)
no_of_unique_bowler=len(df1["bowlerIdx"].unique())
print(no_of_unique_bowler)
embedding_size_bat = no_of_unique_batman ** (1/4)
print(embedding_size_bat)
embedding_size_bwl = no_of_unique_bowler ** (1/4)
print(embedding_size_bwl)
# create embedding layer for the categorical predictor
batsmanIdx_embedding = Embedding(input_dim=no_of_unique_batman+1, output_dim=16,input_length=1)(batsmanIdx_input)
print(batsmanIdx_embedding)
batsmanIdx_flatten = Flatten()(batsmanIdx_embedding)
print(batsmanIdx_flatten)
bowlerIdx_embedding = Embedding(input_dim=no_of_unique_bowler+1, output_dim=16,input_length=1)(bowlerIdx_input)
bowlerIdx_flatten = Flatten()(bowlerIdx_embedding)
print(bowlerIdx_flatten)
# concatenate all the predictors
x = keras.layers.concatenate([batsmanIdx_flatten,bowlerIdx_flatten, ballNum_input, ballsRemaining_input, runs_input, runRate_input, numWickets_input, runsMomentum_input, perfIndex_input])
print(x.shape)
# add hidden layers
x = Dense(96, activation='relu')(x)
x = Dropout(0.1)(x)
x = Dense(64, activation='relu')(x)
x = Dropout(0.1)(x)
x = Dense(32, activation='relu')(x)
x = Dropout(0.1)(x)
x = Dense(16, activation='relu')(x)
x = Dropout(0.1)(x)
x = Dense(8, activation='relu')(x)
x = Dropout(0.1)(x)
# add output layer
output = Dense(1, activation='sigmoid', name='output')(x)
print(output.shape)
# create model
model = Model(inputs=[batsmanIdx_input,bowlerIdx_input, ballNum_input, ballsRemaining_input, runs_input, runRate_input, numWickets_input, runsMomentum_input, perfIndex_input], outputs=output)
model.summary()
# compile model
#optimizer=keras.optimizers.Adam(learning_rate=.01, beta_1=0.1, beta_2=0.999, epsilon=None, decay=0.0, amsgrad=True)
#optimizer=keras.optimizers.RMSprop(learning_rate=0.001, rho=0.2, momentum=0.2, epsilon=1e-07)
#optimizer=keras.optimizers.SGD(learning_rate=.01,momentum=0.1) #- Works without dropout
#optimizer = tf.keras.optimizers.RMSprop(0.01)
#optimizer=keras.optimizers.SGD(learning_rate=.01,momentum=0.1)
#optimizer=keras.optimizers.RMSprop(learning_rate=.005, rho=0.1, momentum=0, epsilon=1e-07)
optimizer=keras.optimizers.Adam(learning_rate=.015, beta_1=0.9, beta_2=0.999, epsilon=1e-07, amsgrad=True)
model.compile(optimizer=optimizer, loss='binary_crossentropy', metrics=['accuracy'])
# train the model
history=model.fit([train_dataset1['batsmanIdx'],train_dataset1['bowlerIdx'],train_dataset1['ballNum'],train_dataset1['ballsRemaining'],train_dataset1['runs'],
train_dataset1['runRate'],train_dataset1['numWickets'],train_dataset1['runsMomentum'],train_dataset1['perfIndex']], train_labels, epochs=20, batch_size=1024,
validation_data = ([test_dataset1['batsmanIdx'],test_dataset1['bowlerIdx'],test_dataset1['ballNum'],test_dataset1['ballsRemaining'],test_dataset1['runs'],
test_dataset1['runRate'],test_dataset1['numWickets'],test_dataset1['runsMomentum'],test_dataset1['perfIndex']],test_labels), verbose=1)
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("model loss")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["train", "test"], loc="upper left")
plt.show()
==================================================================================================
Total params: 144,497
Trainable params: 144,497
Non-trainable params: 0
__________________________________________________________________________________________________
Epoch 1/20
1219/1219 [==============================] - 15s 11ms/step - loss: 0.6285 - accuracy: 0.6372 - val_loss: 0.5164 - val_accuracy: 0.7606
Epoch 2/20
1219/1219 [==============================] - 14s 11ms/step - loss: 0.5594 - accuracy: 0.7121 - val_loss: 0.4920 - val_accuracy: 0.7663
Epoch 3/20
1219/1219 [==============================] - 14s 12ms/step - loss: 0.5338 - accuracy: 0.7244 - val_loss: 0.4541 - val_accuracy: 0.7878
Epoch 4/20
1219/1219 [==============================] - 14s 11ms/step - loss: 0.5176 - accuracy: 0.7317 - val_loss: 0.4226 - val_accuracy: 0.7933
Epoch 5/20
1219/1219 [==============================] - 13s 11ms/step - loss: 0.4966 - accuracy: 0.7420 - val_loss: 0.4547 - val_accuracy: 0.7
...
...
poch 18/20
1219/1219 [==============================] - 14s 11ms/step - loss: 0.4300 - accuracy: 0.7747 - val_loss: 0.3536 - val_accuracy: 0.8288
Epoch 19/20
1219/1219 [==============================] - 14s 12ms/step - loss: 0.4269 - accuracy: 0.7766 - val_loss: 0.3565 - val_accuracy: 0.8302
Epoch 20/20
1219/1219 [==============================] - 14s 11ms/step - loss: 0.4259 - accuracy: 0.7775 - val_loss: 0.3498 - val_accuracy: 0.831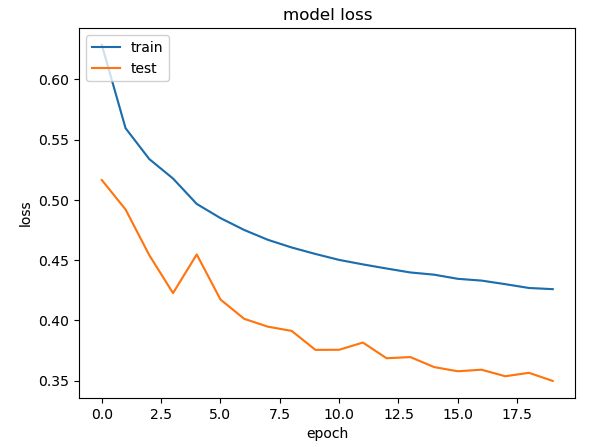
As can be seen the accuracy with augmented dataset is around 0.77, while without it I was getting 0.867 with just the real data. This degradation is probably due to the folllowing reasons
- Only a fraction of the dataset was used for training. This was not representative of the data distribution for CTGAN to correctly synthesise data
- The number of epochs had to be kept low to prevent Kaggle/Colab from crashing
I. Conclusion
This post shows how we can generate synthetic T20 match data to augment real T20 match data. Assuming we have sufficient processing power we should be able to generate synthetic data for augmenting our data set. This should improve the accuracy of the Win Probabily Deep Learning model.
References
- Generative Adversarial Networks – Ian Goodfellow et al.
- Modeling Tabular data using Conditional GAN
- Introduction to GAN
- Ian Goodfellow: Generative Adversarial Networks (GANs) | Lex Fridman Podcast
- CTGAN
- Tabular Synthetic Data Generation using CTGAN
- CTGAN Model
- Interpreting the Progress of CTGAN
- CTGAN metrics
Also see
- Using embeddings, collaborative filtering with Deep Learning to analyse T20 players
- Using Reinforcement Learning to solve Gridworld
- Deep Learning from first principles in Python, R and Octave – Part 4
- Practical Machine Learning with R and Python – Part 5
- Cricketr adds team analytics to its repertoire!!!
- yorkpy takes a hat-trick, bowls out Intl. T20s, BBL and Natwest T20!!!
- Deconstructing Convolutional Neural Networks with Tensorflow and Keras
- My TEDx talk on the “Internet of Things”
- Introducing QCSimulator: A 5-qubit quantum computing simulator in R
- The Anomaly
To see all posts click Index of posts























































 Checkout my book ‘Deep Learning from first principles: Second Edition – In vectorized Python, R and Octave’. My book starts with the implementation of a simple 2-layer Neural Network and works its way to a generic L-Layer Deep Learning Network, with all the bells and whistles. The derivations have been discussed in detail. The code has been extensively commented and included in its entirety in the Appendix sections. My book is available on Amazon as
Checkout my book ‘Deep Learning from first principles: Second Edition – In vectorized Python, R and Octave’. My book starts with the implementation of a simple 2-layer Neural Network and works its way to a generic L-Layer Deep Learning Network, with all the bells and whistles. The derivations have been discussed in detail. The code has been extensively commented and included in its entirety in the Appendix sections. My book is available on Amazon as 








